Development of a Pharmacogenetic Predictive Test in asthma: proof of concept
- PMID: 20032818
- PMCID: PMC3654515
- DOI: 10.1097/FPC.0b013e32833428d0
Development of a Pharmacogenetic Predictive Test in asthma: proof of concept
Abstract
Objective: To assess the feasibility of developing a Combined Clinical and Pharmacogenetic Predictive Test, comprised of multiple single nucleotide polymorphisms (SNPs) that is associated with poor bronchodilator response (BDR).
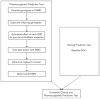
Methods: We genotyped SNPs that tagged the whole genome of the parents and children in the Childhood Asthma Management Program (CAMP) and implemented an algorithm using a family-based association test that ranked SNPs by statistical power. The top eight SNPs that were associated with BDR comprised the Pharmacogenetic Predictive Test. The Clinical Predictive Test was comprised of baseline forced expiratory volume in 1 s (FEV1). We evaluated these predictive tests and a Combined Clinical and Pharmacogenetic Predictive Test in three distinct populations: the children of the CAMP trial and two additional clinical trial populations of asthma. Our outcome measure was poor BDR, defined as BDR of less than 20th percentile in each population. BDR was calculated as the percent difference between the prebronchodilator and postbronchodilator (two puffs of albuterol at 180 microg/puff) FEV1 value. To assess the predictive ability of the test, the corresponding area under the receiver operating characteristic curves (AUROCs) were calculated for each population.
Results: The AUROC values for the Clinical Predictive Test alone were not significantly different from 0.50, the AUROC of a random classifier. Our Combined Clinical and Pharmacogenetic Predictive Test comprised of genetic polymorphisms in addition to FEV1 predicted poor BDR with an AUROC of 0.65 in the CAMP children (n = 422) and 0.60 (n = 475) and 0.63 (n = 235) in the two independent populations. Both the Combined Clinical and Pharmacogenetic Predictive Test and the Pharmacogenetic Predictive Test were significantly more accurate than the Clinical Predictive Test (AUROC between 0.44 and 0.55) in each of the populations.
Conclusion: Our finding that genetic polymorphisms with a clinical trait are associated with BDR suggests that there is promise in using multiple genetic polymorphisms simultaneously to predict which asthmatics are likely to respond poorly to bronchodilators.
Figures

References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural MJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
-
- Christensen K, Murray JC. What genome-wide association studies can do for medicine. N Engl J Med. 2007;356:1094–1097. - PubMed
-
- Hunter DJ, Khoury MJ, Drazen JM. Letting the genome out of the bottle–will we get our wish? N Engl J Med. 2008;358:105–107. - PubMed
-
- Tantisira K, Weiss S. The pharmacogenetics of asthma treatment. Curr Allergy Asthma Rep. 2009;9:10–17. - PubMed
Publication types
MeSH terms
Grants and funding
- HL071394/HL/NHLBI NIH HHS/United States
- P01 HL083069/HL/NHLBI NIH HHS/United States
- HL074755/HL/NHLBI NIH HHS/United States
- R01 HL074755/HL/NHLBI NIH HHS/United States
- U01 HL065899/HL/NHLBI NIH HHS/United States
- K08 HL088046/HL/NHLBI NIH HHS/United States
- U01 HL075419/HL/NHLBI NIH HHS/United States
- K12 HL089990/HL/NHLBI NIH HHS/United States
- R01 HL086601/HL/NHLBI NIH HHS/United States
- T32 HL07427/HL/NHLBI NIH HHS/United States
- T32 HL007427/HL/NHLBI NIH HHS/United States
- U01 HL65899/HL/NHLBI NIH HHS/United States
- R01 HL071394/HL/NHLBI NIH HHS/United States

