The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes
- PMID: 20033039
- PMCID: PMC2869534
- DOI: 10.1038/nature08621
The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes
Abstract
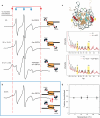
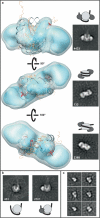
Evenly spaced nucleosomes directly correlate with condensed chromatin and gene silencing. The ATP-dependent chromatin assembly factor (ACF) forms such structures in vitro and is required for silencing in vivo. ACF generates and maintains nucleosome spacing by constantly moving a nucleosome towards the longer flanking DNA faster than the shorter flanking DNA. How the enzyme rapidly moves back and forth between both sides of a nucleosome to accomplish bidirectional movement is unknown. Here we show that nucleosome movement depends cooperatively on two ACF molecules, indicating that ACF functions as a dimer of ATPases. Further, the nucleotide state determines whether the dimer closely engages one or both sides of the nucleosome. Three-dimensional reconstruction by single-particle electron microscopy of the ATPase-nucleosome complex in an activated ATP state reveals a dimer architecture in which the two ATPases face each other. Our results indicate a model in which the two ATPases work in a coordinated manner, taking turns to engage either side of a nucleosome, thereby allowing processive bidirectional movement. This novel dimeric motor mechanism differs from that of dimeric motors such as kinesin and dimeric helicases that processively translocate unidirectionally and reflects the unique challenges faced by motors that move nucleosomes.
Figures


Similar articles
-
Dynamics of nucleosome remodelling by individual ACF complexes.Nature. 2009 Dec 24;462(7276):1022-7. doi: 10.1038/nature08627. Nature. 2009. PMID: 20033040 Free PMC article.
-
The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing.Nat Struct Mol Biol. 2006 Dec;13(12):1078-83. doi: 10.1038/nsmb1170. Epub 2006 Nov 12. Nat Struct Mol Biol. 2006. PMID: 17099699
-
Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.Nature. 2018 Apr;556(7701):386-390. doi: 10.1038/s41586-018-0029-y. Epub 2018 Apr 11. Nature. 2018. PMID: 29643509 Free PMC article.
-
A proposal for kinetic proof reading by ISWI family chromatin remodeling motors.Curr Opin Chem Biol. 2010 Oct;14(5):660-5. doi: 10.1016/j.cbpa.2010.08.001. Epub 2010 Sep 15. Curr Opin Chem Biol. 2010. PMID: 20833099 Free PMC article. Review.
-
Mechanisms for nucleosome movement by ATP-dependent chromatin remodeling complexes.Results Probl Cell Differ. 2006;41:127-48. doi: 10.1007/400_005. Results Probl Cell Differ. 2006. PMID: 16909894 Review.
Cited by
-
The role of the double bromodomain-containing BET genes during mammalian spermatogenesis.Curr Top Dev Biol. 2013;102:293-326. doi: 10.1016/B978-0-12-416024-8.00011-8. Curr Top Dev Biol. 2013. PMID: 23287038 Free PMC article. Review.
-
Sophisticated Conversations between Chromatin and Chromatin Remodelers, and Dissonances in Cancer.Int J Mol Sci. 2021 May 25;22(11):5578. doi: 10.3390/ijms22115578. Int J Mol Sci. 2021. PMID: 34070411 Free PMC article. Review.
-
ISWI remodels nucleosomes through a random walk.Biochemistry. 2014 Jul 15;53(27):4346-57. doi: 10.1021/bi500226b. Epub 2014 Jun 30. Biochemistry. 2014. PMID: 24898619 Free PMC article.
-
Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes.Cell. 2013 Aug 1;154(3):490-503. doi: 10.1016/j.cell.2013.07.011. Cell. 2013. PMID: 23911317 Free PMC article. Review.
-
Structural basis of molecular recognition of helical histone H3 tail by PHD finger domains.Biochem J. 2017 May 4;474(10):1633-1651. doi: 10.1042/BCJ20161053. Biochem J. 2017. PMID: 28341809 Free PMC article.
References
-
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. - PubMed
-
- Varga-Weisz PD, et al. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. - PubMed
-
- Deuring R, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. - PubMed

