A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea
- PMID: 20033048
- PMCID: PMC3073058
- DOI: 10.1038/nature08656
A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea
Abstract
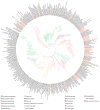
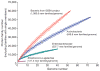
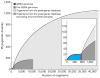
Sequencing of bacterial and archaeal genomes has revolutionized our understanding of the many roles played by microorganisms. There are now nearly 1,000 completed bacterial and archaeal genomes available, most of which were chosen for sequencing on the basis of their physiology. As a result, the perspective provided by the currently available genomes is limited by a highly biased phylogenetic distribution. To explore the value added by choosing microbial genomes for sequencing on the basis of their evolutionary relationships, we have sequenced and analysed the genomes of 56 culturable species of Bacteria and Archaea selected to maximize phylogenetic coverage. Analysis of these genomes demonstrated pronounced benefits (compared to an equivalent set of genomes randomly selected from the existing database) in diverse areas including the reconstruction of phylogenetic history, the discovery of new protein families and biological properties, and the prediction of functions for known genes from other organisms. Our results strongly support the need for systematic 'phylogenomic' efforts to compile a phylogeny-driven 'Genomic Encyclopedia of Bacteria and Archaea' in order to derive maximum knowledge from existing microbial genome data as well as from genome sequences to come.
Figures

Comment in
-
Filling the gaps in the genomic landscape.Genome Biol. 2010;11(2):103. doi: 10.1186/gb-2010-11-2-103. Epub 2010 Feb 16. Genome Biol. 2010. PMID: 20210981 Free PMC article.
References
-
- Eisen JA. Assessing evolutionary relationships among microbes from whole-genome analysis. Curr Opin Microbiol. 2000;3:475–480. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

