Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex
- PMID: 20033062
- PMCID: PMC2829161
- DOI: 10.1038/emboj.2009.386
Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex
Abstract
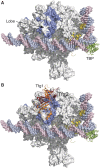
The RNA polymerase (pol) II general transcription factor TFIIF functions at several steps in transcription initiation including preinitiation complex (PIC) formation and start site selection. We find that two structured TFIIF domains bind Pol II at separate locations far from the active site with the TFIIF dimerization domain on the Pol II lobe and the winged helix domain of the TFIIF small subunit Tfg2 above the Pol II protrusion where it may interact with upstream promoter DNA. Binding of the winged helix to the protrusion is PIC specific. Anchoring of these two structured TFIIF domains at separate sites locates an essential and unstructured region of Tfg2 near the Pol II active site cleft where it may interact with flexible regions of Pol II and the general factor TFIIB to promote initiation and start site selection. Consistent with this mechanism, mutations far from the enzyme active site, which alter the binding of either structured TFIIF domains to Pol II, have similar defects in transcription start site usage.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






Similar articles
-
Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry.EMBO J. 2010 Feb 17;29(4):717-26. doi: 10.1038/emboj.2009.401. Epub 2010 Jan 21. EMBO J. 2010. PMID: 20094031 Free PMC article.
-
The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex.Nat Struct Mol Biol. 2007 Aug;14(8):696-703. doi: 10.1038/nsmb1272. Epub 2007 Jul 15. Nat Struct Mol Biol. 2007. PMID: 17632521 Free PMC article.
-
Evidence that the Tfg1/Tfg2 dimer interface of TFIIF lies near the active center of the RNA polymerase II initiation complex.Nucleic Acids Res. 2005 Sep 7;33(16):5045-52. doi: 10.1093/nar/gki825. Print 2005. Nucleic Acids Res. 2005. PMID: 16147988 Free PMC article.
-
RNA polymerase II elongation factors use conserved regulatory mechanisms.Curr Opin Struct Biol. 2024 Feb;84:102766. doi: 10.1016/j.sbi.2023.102766. Epub 2024 Jan 4. Curr Opin Struct Biol. 2024. PMID: 38181687 Review.
-
Structural Insights into the Eukaryotic Transcription Initiation Machinery.Annu Rev Biophys. 2017 May 22;46:59-83. doi: 10.1146/annurev-biophys-070816-033751. Annu Rev Biophys. 2017. PMID: 28532216 Free PMC article. Review.
Cited by
-
Super elongation complex contains a TFIIF-related subcomplex.Transcription. 2016 Aug 7;7(4):133-40. doi: 10.1080/21541264.2016.1194027. Epub 2016 May 25. Transcription. 2016. PMID: 27223670 Free PMC article.
-
Specialization versus conservation: How Pol I and Pol III use the conserved architecture of the pre-initiation complex for specialized transcription.Transcription. 2016 Aug 7;7(4):127-32. doi: 10.1080/21541264.2016.1203628. Epub 2016 Jun 21. Transcription. 2016. PMID: 27327079 Free PMC article. Review.
-
Molecular mechanisms of transcription through single-molecule experiments.Chem Rev. 2014 Mar 26;114(6):3203-23. doi: 10.1021/cr400730x. Epub 2014 Feb 6. Chem Rev. 2014. PMID: 24502198 Free PMC article. Review. No abstract available.
-
Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms.Proc Natl Acad Sci U S A. 2014 May 6;111(18):6642-7. doi: 10.1073/pnas.1405181111. Epub 2014 Apr 14. Proc Natl Acad Sci U S A. 2014. PMID: 24733897 Free PMC article.
-
The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration.Nat Commun. 2014 Oct 14;5:5132. doi: 10.1038/ncomms6132. Nat Commun. 2014. PMID: 25311937 Free PMC article.
References
-
- Bushnell DA, Bamdad C, Kornberg RD (1996) A minimal set of RNA Pol II transcription protein interactions. J Biol Chem 271: 20170–20174 - PubMed
-
- Bushnell DA, Westover KD, Davis RE, Kornberg RD (2004) Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science 303: 983–988 - PubMed
-
- Chen HT, Hahn S (2004) Mapping the location of TFIIB within the RNA Polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119: 169–180 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

