Increased physical activity cosegregates with higher intake of carbohydrate and total calories in a subcongenic mouse strain
- PMID: 20033694
- PMCID: PMC2904081
- DOI: 10.1007/s00335-009-9243-0
Increased physical activity cosegregates with higher intake of carbohydrate and total calories in a subcongenic mouse strain
Abstract
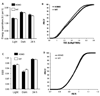
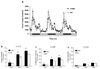
C57BL/6 J (B6) and CAST/EiJ (CAST), the inbred strain derived from M. musculus castaneus, differ in nutrient intake behaviors, including dietary fat and carbohydrate consumption in a two-diet-choice paradigm. Significant quantitative trait loci (QTLs) for carbohydrate (Mnic1) and total energy intake (Kcal2) are present between these strains on chromosome (Chr) 17. Here we report the refinement of the Chr 17 QTL in a subcongenic strain of the B6.CAST-( D17Mit19-D17Mit91 ) congenic mice described previously. This new subcongenic strain possesses CAST Chr 17 donor alleles from 4.8 to 45.4 Mb on a B6 background. Similar to CAST, the subcongenic mice exhibit increased carbohydrate and total calorie intake per body weight, while fat intake remains equivalent. Unexpectedly, this CAST genomic segment also confers two new physical activity phenotypes: 22% higher spontaneous physical activity levels and significantly increased voluntary wheel-running activity compared with the parental B6 strain. Overall, these data suggest that gene(s) involved in carbohydrate preference and increased physical activity are contained within the proximal region of Chr 17. Interval-specific microarray analysis in hypothalamus and skeletal muscle revealed differentially expressed genes within the subcongenic region, including neuropeptide W (Npw); glyoxalase I (Glo1); cytochrome P450, family 4, subfamily f, polypeptide 1 (Cyp4f15); phospholipase A2, group VII (Pla2g7); and phosphodiesterase 9a (Pde9a). This subcongenic strain offers a unique model for dissecting the contributions and possible interactions among genes controlling food intake and physical activity, key components of energy balance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Albarado DC, McClaine J, Stephens JM, Mynatt RL, Ye J, et al. Impaired coordination of nutrient intake and substrate oxidation in melanocortin-4 receptor knockout mice. Endocrinol. 2004;145:243–252. - PubMed
-
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
-
- Cai G, Cole SA, Bastarrachea RA, Maccluer JW, Blangero J, et al. Quantitative trait locus determining dietary macronutrient intakes is located on human chromosome 2p22. Am J Clin Nutr. 2004;80:1410–1414. - PubMed
-
- Cai G, Cole SA, Butte N, Bacino C, Diego V, et al. A quantitative trait locus on chromosome 18q for physical activity and dietary intake in Hispanic children. Obesity (Silver Spring) 2006;14:1596–1604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

