Transcriptional regulatory regions of gap43 needed in developing and regenerating retinal ganglion cells
- PMID: 20034105
- PMCID: PMC3101714
- DOI: 10.1002/dvdy.22190
Transcriptional regulatory regions of gap43 needed in developing and regenerating retinal ganglion cells
Abstract
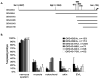
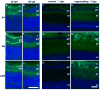
Mammals and fish differ in their ability to express axon growth-associated genes in response to CNS injury, which contributes to the differences in their ability for CNS regeneration. Previously we demonstrated that for the axon growth-associated gene, gap43, regions of the rat promoter that are sufficient to promote reporter gene expression in the developing zebrafish nervous system are not sufficient to promote expression in regenerating retinal ganglion cells in zebrafish. Recently, we identified a 3.6-kb gap43 promoter fragment from the pufferfish, Takifugu rubripes (fugu), that can promote reporter gene expression during both development and regeneration. Using promoter deletion analysis, we have found regions of the 3.6-kb fugu gap43 promoter that are necessary for expression in regenerating, but not developing, retinal ganglion cells. Within the 3.6-kb promoter, we have identified elements that are highly conserved among fish, as well as elements conserved among fish, mammals, and birds.
Figures




Similar articles
-
3.6 kb genomic sequence from Takifugu capable of promoting axon growth-associated gene expression in developing and regenerating zebrafish neurons.Gene Expr Patterns. 2008 Jul;8(6):382-388. doi: 10.1016/j.gep.2008.05.002. Epub 2008 May 24. Gene Expr Patterns. 2008. PMID: 18599366 Free PMC article.
-
An element in the alpha1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish.J Neurosci. 2004 Sep 1;24(35):7663-73. doi: 10.1523/JNEUROSCI.2281-04.2004. J Neurosci. 2004. PMID: 15342733 Free PMC article.
-
GAP-43 promoter elements in transgenic zebrafish reveal a difference in signals for axon growth during CNS development and regeneration.Development. 2001 Apr;128(7):1175-82. doi: 10.1242/dev.128.7.1175. Development. 2001. PMID: 11245583
-
Retina regeneration in zebrafish.Curr Opin Genet Dev. 2016 Oct;40:41-47. doi: 10.1016/j.gde.2016.05.009. Epub 2016 Jun 6. Curr Opin Genet Dev. 2016. PMID: 27281280 Free PMC article. Review.
-
A molecular mechanism of optic nerve regeneration in fish: the retinoid signaling pathway.Prog Retin Eye Res. 2013 Nov;37:13-30. doi: 10.1016/j.preteyeres.2013.07.004. Epub 2013 Aug 28. Prog Retin Eye Res. 2013. PMID: 23994437 Review.
Cited by
-
Extrinsic and intrinsic determinants of nerve regeneration.J Tissue Eng. 2011;2(1):2041731411418392. doi: 10.1177/2041731411418392. Epub 2011 Sep 13. J Tissue Eng. 2011. PMID: 22292105 Free PMC article.
-
Deletion of Slc1a4 Suppresses Single Mauthner Cell Axon Regeneration In Vivo through Growth-Associated Protein 43.Int J Mol Sci. 2024 Oct 11;25(20):10950. doi: 10.3390/ijms252010950. Int J Mol Sci. 2024. PMID: 39456733 Free PMC article.
-
Acute myocardial infarction induces remodeling of the murine superior cervical ganglia and the carotid body.Front Cardiovasc Med. 2022 Oct 6;9:758265. doi: 10.3389/fcvm.2022.758265. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36277772 Free PMC article.
-
MASH1/Ascl1a leads to GAP43 expression and axon regeneration in the adult CNS.PLoS One. 2015 Mar 9;10(3):e0118918. doi: 10.1371/journal.pone.0118918. eCollection 2015. PLoS One. 2015. PMID: 25751153 Free PMC article.
-
Serotonin neuromodulation directs optic nerve regeneration.Development. 2025 Jul 1;152(13):dev204334. doi: 10.1242/dev.204334. Epub 2025 Jul 7. Development. 2025. PMID: 40521655 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Barthel LK, Raymond PA. Improved method for obtaining 3-microns cryosections for immunocytochemistry. J Histochem Cytochem. 1990;38:1383–1388. - PubMed
-
- Becker CG, Becker T. Growth and pathfinding of regenerating axons in the optic projection of adult fish. J Neurosci Res. 2007;85:2793–2799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

