Next generation transcriptomes for next generation genomes using est2assembly
- PMID: 20034392
- PMCID: PMC3087352
- DOI: 10.1186/1471-2105-10-447
Next generation transcriptomes for next generation genomes using est2assembly
Abstract
Background: The decreasing costs of capillary-based Sanger sequencing and next generation technologies, such as 454 pyrosequencing, have prompted an explosion of transcriptome projects in non-model species, where even shallow sequencing of transcriptomes can now be used to examine a range of research questions. This rapid growth in data has outstripped the ability of researchers working on non-model species to analyze and mine transcriptome data efficiently.
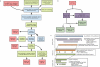
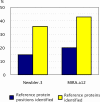
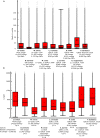
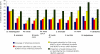
Results: Here we present a semi-automated platform 'est2assembly' that processes raw sequence data from Sanger or 454 sequencing into a hybrid de-novo assembly, annotates it and produces GMOD compatible output, including a SeqFeature database suitable for GBrowse. Users are able to parameterize assembler variables, judge assembly quality and determine the optimal assembly for their specific needs. We used est2assembly to process Drosophila and Bicyclus public Sanger EST data and then compared them to published 454 data as well as eight new insect transcriptome collections.
Conclusions: Analysis of such a wide variety of data allows us to understand how these new technologies can assist EST project design. We determine that assembler parameterization is as essential as standardized methods to judge the output of ESTs projects. Further, even shallow sequencing using 454 produces sufficient data to be of wide use to the community. est2assembly is an important tool to assist manual curation for gene models, an important resource in their own right but especially for species which are due to acquire a genome project using Next Generation Sequencing.
Figures


References
-
- Van Straalen NM, Roelofs D. An introduction to ecological genomics. Oxford: Oxford University Press; 2006.
-
- Heckel DG, Gahan LJ, Daly JC, Trowell S. A genomic approach to understanding Heliothis and Helicoverpa resistance to chemical and biological insecticides. Philos Trans R Soc Lond B Biol Sci. 1998;353:1713–1722.
-
- Brakefield PM, Gates J, Keys D, Kesbeke F, Wijngaarden PJ, Monteiro A, French V, Carroll SB. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 1996;384:236–242. - PubMed
-
- Rausher MD. In: Insect chemical ecology: an evolutionary approach. Rausher MD, Isman MB, editor. New York: Chapman & Hall; 1992. Natural selection and the evolution of plant insect interactions; pp. 20–88.
-
- Ewing B, Green P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet. 2000;25:232–234. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

