Chitosan-modified poly(D,L-lactide-co-glycolide) nanospheres for improving siRNA delivery and gene-silencing effects
- PMID: 20034563
- PMCID: PMC7127408
- DOI: 10.1016/j.ejpb.2009.12.007
Chitosan-modified poly(D,L-lactide-co-glycolide) nanospheres for improving siRNA delivery and gene-silencing effects
Abstract
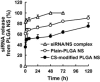
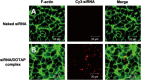
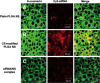
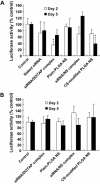
Chitosan (CS) surface-modified poly(D,L-lactide-co-glycolide) (PLGA) nanospheres (NS) for a siRNA delivery system were evaluated in vitro. siRNA-loaded PLGA NS were prepared by an emulsion solvent diffusion (ESD) method, and the physicochemical properties of NS were investigated. The level of targeted protein expression and siRNA uptake were examined in A549 cells. CS-modified PLGA NS exhibited much higher encapsulation efficiency than unmodified PLGA NS (plain-PLGA NS). CS-modified PLGA NS showed a positive zeta potential, while plain-PLGA NS were negatively charged. siRNA uptake studies by observation with confocal laser scanning microscopy (CLSM) indicated that siRNA-loaded CS-modified PLGA NS were more effectively taken up by the cells than plain-PLGA NS. The efficiencies of different siRNA preparations were compared at the level of targeted protein expression. The gene-silencing efficiency of CS-modified PLGA NS was higher and more prolonged than those of plain-PLGA NS and naked siRNA. This result correlated with the CLSM studies, which may have been due to higher cellular uptake of CS-modified PLGA NS due to electrostatic interactions. It was concluded that CS-modified PLGA NS containing siRNA could provide an effective siRNA delivery system.
(c) 2009 Elsevier B.V. All rights reserved.
Figures

References
-
- Yano J., Hirabayashi K., Nakagawa S., Yamaguchi T., Nogawa M., Kashimori I., Naito H., Kitagawa H., Ishiyama K., Ohgi T., Irimura T. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin. Cancer Res. 2004;10:7721–7726. - PubMed
-
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. - PubMed
-
- Smith J., Zhang Y., Niven R. Toward development of a nonviral gene therapeutic. Adv. Drug. Deliv. Rev. 1997;26:135–150. - PubMed
-
- Büning H., Nicklin S.A., Perabo L., Hallek M., Baker A.H. AAV-based gene transfer. Curr. Opin. Mol. Ther. 2003;5:367–375. - PubMed
-
- Rudolph C., Plank C., Lausier J., Schillinger U., Müller R.H., Rosenecker J. Oligomers of the arginine-rich motif of the HIV-1 TAT protein are capable of transferring plasmid DNA into cells. J. Biol. Chem. 2003;278:11411–11418. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

