A time-resolved fluorescence-resonance energy transfer assay for identifying inhibitors of hepatitis C virus core dimerization
- PMID: 20035614
- PMCID: PMC2971647
- DOI: 10.1089/adt.2009.0217
A time-resolved fluorescence-resonance energy transfer assay for identifying inhibitors of hepatitis C virus core dimerization
Abstract
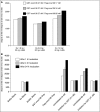
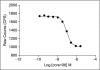
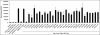
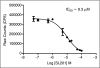
Binding of hepatitis C virus (HCV) RNA to core, the capsid protein, results in the formation of the nucleocapsid, the first step in the assembly of the viral particle. A novel assay was developed to discover small molecule inhibitors of core dimerization. This assay is based on time-resolved fluorescence resonance energy transfer (TR-FRET) between anti-tag antibodies labeled with either europium cryptate (Eu) or allophycocyanin (XL-665). The N-terminal 106-residue portion of core protein (core106) was tagged with either glutathione-S-transferase (GST) or a Flag peptide. Tag-free core106 was selected as the reference inhibitor. The assay was used to screen the library of pharmacologically active compounds (LOPAC) consisting of 1,280 compounds and a 2,240-compound library from the Center for Chemical Methodology and Library Development at Boston University (CMLD-BU). Ten of the 28 hits from the primary TR-FRET run were confirmed in a secondary amplified luminescent proximity homogeneous assay (ALPHA screen). One hit was further characterized by dose-response analysis yielding an IC(50) of 9.3 microM. This 513 Da compound was shown to inhibit HCV production in cultured hepatoma cells.
Figures

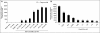

References
-
- WHO. Hepatitis C: global prevalence. Wkly Epidemiol Rec. 1997;72:341–344. - PubMed
-
- Giannini C. Brechot C. Hepatitis C virus biology. Cell Death Differ. 2003;10(Suppl. 1):S27–S38. - PubMed
-
- Choo Q. Kuo G. Weiner A. Overby L. Bradley D. Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359. - PubMed
-
- McGovern BH. Hepatitis C in the HIV-infected patient. J Acquir Immune Defic Syndr. 2007;45(Suppl 2):S47–S56. - PubMed
-
- Cristina J. Moreno-del Pilar M. Moratorio G. Hepatitis C virus genetic variability in patients undergoing antiviral therapy. Virus Res. 2007;127(2):185–194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

