Non-natural amino acid fluorophores for one- and two-step fluorescence resonance energy transfer applications
- PMID: 20036210
- PMCID: PMC2830288
- DOI: 10.1016/j.ab.2009.12.027
Non-natural amino acid fluorophores for one- and two-step fluorescence resonance energy transfer applications
Abstract
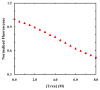
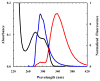
Fluorescence resonance energy transfer (FRET) provides a powerful means to study protein conformational changes. However, the incorporation of an exogenous FRET pair into a protein could lead to undesirable structural perturbations of the native fold. One of the viable strategies to minimizing such perturbations is to use non-natural amino acid-based FRET pairs. Previously, we showed that p-cyanophenylalanine (Phe(CN)) and tryptophan (Trp) constitute such a FRET pair, useful for monitoring protein folding-unfolding transitions. Here we further show that 7-azatryptophan (7AW) and 5-hydroxytryptophan (5HW) can also serve as a FRET acceptor to Phe(CN), and the resultant FRET pairs offer certain advantages over Phe(CN)-Trp. For example, the fluorescence spectrum of 7AW is sufficiently separated from that of Phe(CN), making it straightforward to decompose the FRET spectrum into donor and acceptor contributions. Moreover, we show that Phe(CN), Trp, and 7AW can be used together to form a multi-FRET system, allowing more structural information to be extracted from a single FRET experiment. The applicability of these FRET systems is demonstrated in a series of studies where they are employed to monitor the urea-induced unfolding transitions of the villin headpiece subdomain (HP35), a designed betabetaalpha motif (BBA5), and the human Pin1 WW domain.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures







Similar articles
-
Using an amino acid fluorescence resonance energy transfer pair to probe protein unfolding: application to the villin headpiece subdomain and the LysM domain.Biochemistry. 2008 Oct 21;47(42):11070-6. doi: 10.1021/bi8012406. Epub 2008 Sep 25. Biochemistry. 2008. PMID: 18816063 Free PMC article.
-
Native state conformational heterogeneity of HP35 revealed by time-resolved FRET.J Phys Chem B. 2012 Sep 6;116(35):10631-8. doi: 10.1021/jp211296e. Epub 2012 Aug 27. J Phys Chem B. 2012. PMID: 22891809 Free PMC article.
-
Characterization of aminoacyl-adenylates in B. subtilis tryptophanyl-tRNA synthetase, by the fluorescence of tryptophan analogs 5-hydroxytryptophan and 7-azatryptophan.Biophys Chem. 1993 Dec;48(2):159-69. doi: 10.1016/0301-4622(93)85007-5. Biophys Chem. 1993. PMID: 8298054
-
Multi-fluorophore fluorescence resonance energy transfer for probing nucleic acids structure and folding.Methods Mol Biol. 2006;335:257-71. doi: 10.1385/1-59745-069-3:257. Methods Mol Biol. 2006. PMID: 16785633 Review.
-
Förster resonance energy transfer and kinesin motor proteins.Chem Soc Rev. 2014 Feb 21;43(4):1144-55. doi: 10.1039/c3cs60292c. Chem Soc Rev. 2014. PMID: 24071719 Review.
Cited by
-
β-Glutamine-mediated self-association of transmembrane β-peptides within lipid bilayers.Chem Sci. 2016 Sep 1;7(9):5900-5907. doi: 10.1039/c6sc01147k. Epub 2016 May 19. Chem Sci. 2016. PMID: 30034732 Free PMC article.
-
Site-Specific Spectroscopic Reporters of the Local Electric Field, Hydration, Structure, and Dynamics of Biomolecules.J Phys Chem Lett. 2011 Sep 23;2:2598-2609. doi: 10.1021/jz201161b. J Phys Chem Lett. 2011. PMID: 22003429 Free PMC article.
-
Modulation of p-cyanophenylalanine fluorescence by amino acid side chains and rational design of fluorescence probes of alpha-helix formation.Biochemistry. 2010 Jul 27;49(29):6290-5. doi: 10.1021/bi100932p. Biochemistry. 2010. PMID: 20565125 Free PMC article.
-
Site-Specific Interrogation of Protein Structure and Stability.Methods Mol Biol. 2022;2376:65-87. doi: 10.1007/978-1-0716-1716-8_3. Methods Mol Biol. 2022. PMID: 34845603
-
Meandering Down the Energy Landscape of Protein Folding: Are We There Yet?Biophys J. 2016 May 10;110(9):1924-32. doi: 10.1016/j.bpj.2016.03.030. Biophys J. 2016. PMID: 27166801 Free PMC article. Review.
References
-
-
Förster T. Annalen der Physik. 1948;2:55–75.; English translation in Mielczarek EV, Greenbaum E, Knox RS, editors. Biological Physics. American Institute of Physics; New York: 1993. pp. 148–160.
-
-
- Hass E. The study of protein folding and dynamics by determination of intermolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements. ChemPhysChem. 2005;6:858–870. - PubMed
-
- Tucker MJ, Oyola R, Gai F. Conformational distribution of a 14-residue peptide in solution: A fluorescence resonance energy transfer study. J Phys Chem B. 2005;109:4788–4795. - PubMed
-
- Tucker MJ, Tang J, Gai F. Probing the kinetics of membrane-mediated helix folding. J Phys Chem B. 2006;110:8105–8109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

