Role of transmembrane domain 4 in ligand permeation by Crithidia fasciculata equilibrative nucleoside transporter 2 (CfNT2)
- PMID: 20037157
- PMCID: PMC2825396
- DOI: 10.1074/jbc.M109.074351
Role of transmembrane domain 4 in ligand permeation by Crithidia fasciculata equilibrative nucleoside transporter 2 (CfNT2)
Abstract
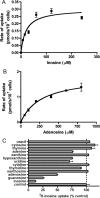
Equilibrative nucleoside transporters play essential roles in nutrient uptake, cardiovascular and renal function, and purine analog drug chemotherapies. Limited structural information is available for this family of transporters; however, residues in transmembrane domains 1, 2, 4, and 5 appear to be important for ligand and inhibitor binding. In order to identify regions of the transporter that are important for ligand specificity, a genetic selection for mutants of the inosine-guanosine-specific Crithidia fasciculata nucleoside transporter 2 (CfNT2) that had gained the ability to transport adenosine was carried out in the yeast Saccharomyces cerevisiae. Nearly all positive clones from the genetic selection carried mutations at lysine 155 in transmembrane domain 4, highlighting lysine 155 as a pivotal residue governing the ligand specificity of CfNT2. Mutation of lysine 155 to asparagine conferred affinity for adenosine on the mutant transporter at the expense of inosine and guanosine affinity due to weakened contacts to the purine ring of the ligand. Following systematic cysteine-scanning mutagenesis, thiol-specific modification of several positions within transmembrane domain 4 was found to interfere with inosine transport capability, indicating that this helix lines the water-filled ligand translocation channel. Additionally, the pattern of modification of transmembrane domain 4 suggested that it may deviate from helicity in the vicinity of residue 155. Position 155 was also protected from modification in the presence of ligand, suggesting that lysine 155 is in or near the ligand binding site. Transmembrane domain 4 and particularly lysine 155 appear to play key roles in ligand discrimination and translocation by CfNT2.
Figures




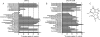
References
-
- Hyde R. J., Cass C. E., Young J. D., Baldwin S. A. (2001) Mol. Membr. Biol. 18, 53–63 - PubMed
-
- Elwi A. N., Damaraju V. L., Baldwin S. A., Young J. D., Sawyer M. B., Cass C. E. (2006) Biochem. Cell Biol. 84, 844–858 - PubMed
-
- Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. (2008) Xenobiotica 38, 995–1021 - PubMed
-
- Molina-Arcas M., Trigueros-Motos L., Casado F. J., Pastor-Anglada M. (2008) Nucleosides Nucleotides Nucleic Acids 27, 769–778 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

