Abrogation of glucocorticoid receptor dimerization correlates with dissociated glucocorticoid behavior of compound a
- PMID: 20037160
- PMCID: PMC2832957
- DOI: 10.1074/jbc.M109.087866
Abrogation of glucocorticoid receptor dimerization correlates with dissociated glucocorticoid behavior of compound a
Abstract
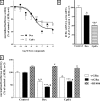
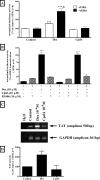
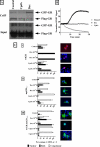
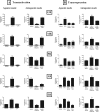
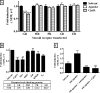
Compound A (CpdA), a dissociated glucocorticoid receptor modulator, decreases corticosteroid-binding globulin (CBG), adrenocorticotropic hormone (ACTH), and luteneinizing hormone levels in rats. Whether this is due to transcriptional regulation by CpdA is not known. Using promoter reporter assays we show that CpdA, like dexamethasone (Dex), directly transrepresses these genes. Results using a rat Cbg proximal-promoter reporter construct in BWTG3 and HepG2 cell lines support a glucocorticoid receptor (GR)-dependent transrepression mechanism for CpdA. However, CpdA, unlike Dex, does not result in transactivation via glucocorticoid-responsive elements within a promoter reporter construct even when GR is co-transfected. The inability of CpdA to result in transactivation via glucocorticoid-responsive elements is confirmed on the endogenous tyrosine aminotransferase gene, whereas transrepression ability is confirmed on the endogenous CBG gene. Consistent with a role for CpdA in modulating GR activity, whole cell binding assays revealed that CpdA binds reversibly to the GR, but with lower affinity than Dex, and influences association of [(3)H]Dex, but has no effect on dissociation. In addition, like Dex, CpdA causes nuclear translocation of the GR, albeit to a lesser degree. Several lines of evidence, including fluorescence resonance energy transfer, co-immunoprecipitation, and nuclear immunofluorescence studies of nuclear localization-deficient GR show that CpdA, unlike Dex, does not elicit ligand-induced GR dimerization. Comparison of the behavior of CpdA in the presence of wild type GR to that of Dex with a dimerization-deficient GR mutant (GR(dim)) strongly supports the conclusion that loss of dimerization is responsible for the dissociated behavior of CpdA.
Figures
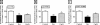


Similar articles
-
Selective modulation of the glucocorticoid receptor can distinguish between transrepression of NF-κB and AP-1.Cell Mol Life Sci. 2014 Jan;71(1):143-63. doi: 10.1007/s00018-013-1367-4. Epub 2013 Jun 20. Cell Mol Life Sci. 2014. PMID: 23784308 Free PMC article.
-
A structural explanation of the effects of dissociated glucocorticoids on glucocorticoid receptor transactivation.Mol Pharmacol. 2014 Feb;85(2):226-36. doi: 10.1124/mol.113.085860. Epub 2013 Nov 13. Mol Pharmacol. 2014. PMID: 24225022
-
A dissociated glucocorticoid receptor modulator reduces airway hyperresponsiveness and inflammation in a mouse model of asthma.J Immunol. 2012 Apr 1;188(7):3478-87. doi: 10.4049/jimmunol.1004227. Epub 2012 Mar 5. J Immunol. 2012. PMID: 22393156
-
New insights into the anti-inflammatory mechanisms of glucocorticoids: an emerging role for glucocorticoid-receptor-mediated transactivation.Endocrinology. 2013 Mar;154(3):993-1007. doi: 10.1210/en.2012-2045. Epub 2013 Feb 5. Endocrinology. 2013. PMID: 23384835 Review.
-
Separating transrepression and transactivation: a distressing divorce for the glucocorticoid receptor?Mol Pharmacol. 2007 Oct;72(4):799-809. doi: 10.1124/mol.107.038794. Epub 2007 Jul 10. Mol Pharmacol. 2007. PMID: 17622575 Review.
Cited by
-
Glucocorticoids suppress inflammation via the upregulation of negative regulator IRAK-M.Nat Commun. 2015 Jan 14;6:6062. doi: 10.1038/ncomms7062. Nat Commun. 2015. PMID: 25585690 Free PMC article.
-
Org 214007-0: a novel non-steroidal selective glucocorticoid receptor modulator with full anti-inflammatory properties and improved therapeutic index.PLoS One. 2012;7(11):e48385. doi: 10.1371/journal.pone.0048385. Epub 2012 Nov 12. PLoS One. 2012. PMID: 23152771 Free PMC article.
-
Noise trauma and systemic application of the selective glucocorticoid receptor modulator compound A.J Negat Results Biomed. 2016 May 11;15:10. doi: 10.1186/s12952-016-0053-0. J Negat Results Biomed. 2016. PMID: 27164957 Free PMC article.
-
Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor.PLoS Biol. 2014 Mar 18;12(3):e1001813. doi: 10.1371/journal.pbio.1001813. eCollection 2014 Mar. PLoS Biol. 2014. PMID: 24642507 Free PMC article.
-
Compound A, a dissociated glucocorticoid receptor modulator, inhibits T-bet (Th1) and induces GATA-3 (Th2) activity in immune cells.PLoS One. 2012;7(4):e35155. doi: 10.1371/journal.pone.0035155. Epub 2012 Apr 9. PLoS One. 2012. PMID: 22496903 Free PMC article.
References
-
- Swart P., Swart A. C., Louw A., van der Merwe K. J. (2003) Bioessays 25, 612–619 - PubMed
-
- Louw A., Swart P., de Kock S. S., van der Merwe K. J. (1997) Biochem. Pharmacol. 53, 189–197 - PubMed
-
- Louw A., Allie F., Swart A. C., Swart P. (2000) Endocr. Res. 26, 729–736 - PubMed
-
- Louw A., Swart P., Allie F. (2000) Biochem. Pharmacol. 59, 167–175 - PubMed
-
- Louw A., Swart P. (1999) Endocrinology 140, 2044–2053 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

