Use of bacterially expressed dsRNA to downregulate Entamoeba histolytica gene expression
- PMID: 20037645
- PMCID: PMC2793006
- DOI: 10.1371/journal.pone.0008424
Use of bacterially expressed dsRNA to downregulate Entamoeba histolytica gene expression
Abstract
Background: Modern RNA interference (RNAi) methodologies using small interfering RNA (siRNA) oligonucleotide duplexes or episomally synthesized hairpin RNA are valuable tools for the analysis of gene function in the protozoan parasite Entamoeba histolytica. However, these approaches still require time-consuming procedures including transfection and drug selection, or costly synthetic molecules.
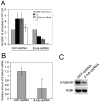
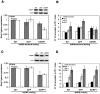
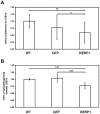
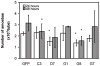
Principal findings: Here we report an efficient and handy alternative for E. histolytica gene down-regulation mediated by bacterial double-stranded RNA (dsRNA) targeting parasite genes. The Escherichia coli strain HT115 which is unable to degrade dsRNA, was genetically engineered to produce high quantities of long dsRNA segments targeting the genes that encode E. histolytica beta-tubulin and virulence factor KERP1. Trophozoites cultured in vitro were directly fed with dsRNA-expressing bacteria or soaked with purified dsRNA. Both dsRNA delivery methods resulted in significant reduction of protein expression. In vitro host cell-parasite assays showed that efficient downregulation of kerp1 gene expression mediated by bacterial dsRNA resulted in significant reduction of parasite adhesion and lytic capabilities, thus supporting a major role for KERP1 in the pathogenic process. Furthermore, treatment of trophozoites cultured in microtiter plates, with a repertoire of eighty-five distinct bacterial dsRNA segments targeting E. histolytica genes with unknown function, led to the identification of three genes potentially involved in the growth of the parasite.
Conclusions: Our results showed that the use of bacterial dsRNA is a powerful method for the study of gene function in E. histolytica. This dsRNA delivery method is also technically suitable for the study of a large number of genes, thus opening interesting perspectives for the identification of novel drug and vaccine targets.
Conflict of interest statement
Figures

Similar articles
-
Silencing the cleavage factor CFIm25 as a new strategy to control Entamoeba histolytica parasite.J Microbiol. 2017 Oct;55(10):783-791. doi: 10.1007/s12275-017-7259-9. Epub 2017 Sep 28. J Microbiol. 2017. PMID: 28956353
-
Double-stranded RNA mediates homology-dependent gene silencing of gamma-tubulin in the human parasite Entamoeba histolytica.Mol Biochem Parasitol. 2004 Nov;138(1):21-8. doi: 10.1016/j.molbiopara.2004.07.005. Mol Biochem Parasitol. 2004. PMID: 15500912
-
Loss of dsRNA-based gene silencing in Entamoeba histolytica: implications for approaches to genetic analysis.Exp Parasitol. 2008 Jun;119(2):296-300. doi: 10.1016/j.exppara.2008.02.001. Epub 2008 Feb 9. Exp Parasitol. 2008. PMID: 18346737 Free PMC article.
-
RNA interference in Entamoeba histolytica: implications for parasite biology and gene silencing.Future Microbiol. 2011 Jan;6(1):103-17. doi: 10.2217/fmb.10.154. Future Microbiol. 2011. PMID: 21162639 Free PMC article. Review.
-
Iron-modulated virulence factors of Entamoeba histolytica.Future Microbiol. 2018 Sep;13:1329-1341. doi: 10.2217/fmb-2018-0066. Epub 2018 Sep 21. Future Microbiol. 2018. PMID: 30238768 Review.
Cited by
-
Targeting the polyadenylation factor EhCFIm25 with RNA aptamers controls survival in Entamoeba histolytica.Sci Rep. 2018 Apr 9;8(1):5720. doi: 10.1038/s41598-018-23997-w. Sci Rep. 2018. PMID: 29632392 Free PMC article.
-
Tissue destruction and invasion by Entamoeba histolytica.Trends Parasitol. 2011 Jun;27(6):254-63. doi: 10.1016/j.pt.2011.02.006. Epub 2011 Mar 26. Trends Parasitol. 2011. PMID: 21440507 Free PMC article. Review.
-
EhVps35, a retromer component, is a key factor in secretion, motility, and tissue invasion by Entamoeba histolytica.Front Cell Infect Microbiol. 2024 Sep 27;14:1467440. doi: 10.3389/fcimb.2024.1467440. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39397861 Free PMC article.
-
Protein Sumoylation Is Crucial for Phagocytosis in Entamoeba histolytica Trophozoites.Int J Mol Sci. 2021 May 27;22(11):5709. doi: 10.3390/ijms22115709. Int J Mol Sci. 2021. PMID: 34071922 Free PMC article.
-
Robust gene silencing mediated by antisense small RNAs in the pathogenic protist Entamoeba histolytica.Nucleic Acids Res. 2013 Nov;41(20):9424-37. doi: 10.1093/nar/gkt717. Epub 2013 Aug 9. Nucleic Acids Res. 2013. PMID: 23935116 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

