Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics
- PMID: 20038105
- PMCID: PMC2834293
- DOI: 10.1021/jm9012455
Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics
Abstract
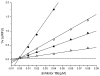
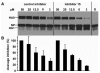
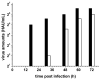
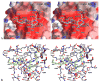
Furin belongs to the family of proprotein convertases (PCs) and is involved in numerous normal physiological and pathogenic processes, such as viral propagation, bacterial toxin activation, cancer, and metastasis. Furin and related furin-like PCs cleave their substrates at characteristic multibasic consensus sequences, preferentially after an arginine residue. By incorporating decarboxylated arginine mimetics in the P1 position of substrate analogue peptidic inhibitors, we could identify highly potent furin inhibitors. The most potent compound, phenylacetyl-Arg-Val-Arg-4-amidinobenzylamide (15), inhibits furin with a K(i) value of 0.81 nM and has also comparable affinity to other PCs like PC1/3, PACE4, and PC5/6, whereas PC2 and PC7 or trypsin-like serine proteases were poorly affected. In fowl plague virus (influenza A, H7N1)-infected MDCK cells, inhibitor 15 inhibited proteolytic hemagglutinin cleavage and was able to reduce virus propagation in a long-term infection test. Molecular modeling revealed several key interactions of the 4-amidinobenzylamide residue in the S1 pocket of furin contributing to the excellent affinity of these inhibitors.
Figures
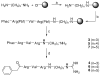

Similar articles
-
Highly potent inhibitors of proprotein convertase furin as potential drugs for treatment of infectious diseases.J Biol Chem. 2012 Jun 22;287(26):21992-2003. doi: 10.1074/jbc.M111.332643. Epub 2012 Apr 26. J Biol Chem. 2012. PMID: 22539349 Free PMC article.
-
Comparative study of the binding pockets of mammalian proprotein convertases and its implications for the design of specific small molecule inhibitors.Int J Biol Sci. 2010 Feb 3;6(1):89-95. doi: 10.7150/ijbs.6.89. Int J Biol Sci. 2010. PMID: 20151049 Free PMC article.
-
Barley serine proteinase inhibitor 2-derived cyclic peptides as potent and selective inhibitors of convertases PC1/3 and furin.Biochemistry. 2003 Aug 19;42(32):9659-68. doi: 10.1021/bi034418w. Biochemistry. 2003. PMID: 12911307
-
[Structure and properties of proprotein convertase inhibitors].Ukr Biokhim Zh (1999). 2012 Mar-Apr;84(2):5-29. Ukr Biokhim Zh (1999). 2012. PMID: 22642118 Review. Russian.
-
The proprotein convertases furin and PACE4 play a significant role in tumor progression.Mol Carcinog. 2000 Jun;28(2):63-9. Mol Carcinog. 2000. PMID: 10900462 Review.
Cited by
-
PACE4 inhibitors and their peptidomimetic analogs block prostate cancer tumor progression through quiescence induction, increased apoptosis and impaired neovascularisation.Oncotarget. 2015 Feb 28;6(6):3680-93. doi: 10.18632/oncotarget.2918. Oncotarget. 2015. PMID: 25682874 Free PMC article.
-
Inhibition of prohormone convertases PC1/3 and PC2 by 2,5-dideoxystreptamine derivatives.Mol Pharmacol. 2012 Mar;81(3):440-54. doi: 10.1124/mol.111.077040. Epub 2011 Dec 14. Mol Pharmacol. 2012. PMID: 22169851 Free PMC article.
-
Furin-mediated protein processing in infectious diseases and cancer.Clin Transl Immunology. 2019 Aug 5;8(8):e1073. doi: 10.1002/cti2.1073. eCollection 2019. Clin Transl Immunology. 2019. PMID: 31406574 Free PMC article. Review.
-
Functional Roles of Furin in Cardio-Cerebrovascular Diseases.ACS Pharmacol Transl Sci. 2024 Feb 7;7(3):570-585. doi: 10.1021/acsptsci.3c00325. eCollection 2024 Mar 8. ACS Pharmacol Transl Sci. 2024. PMID: 38481703 Free PMC article. Review.
-
In vitro characterization of the furin inhibitor MI-1851: Albumin binding, interaction with cytochrome P450 enzymes and cytotoxicity.Biomed Pharmacother. 2022 Jul;151:113124. doi: 10.1016/j.biopha.2022.113124. Epub 2022 May 17. Biomed Pharmacother. 2022. PMID: 35594709 Free PMC article.
References
-
- van de Ven WJ, Voorberg J, Fontijn R, Pannekoek H, van den Ouweland AM, van Duijnhoven HL, Roebroek AJ, Siezen RJ. Furin is a subtilisin-like proprotein processing enzyme in higher eukaryotes. Mol Biol Rep. 1990;14:265–275. - PubMed
-
- Rockwell NC, Krysan DJ, Komiyama T, Fuller RS. Precursor processing by kex2/furin proteases. Chem Rev. 2002;102:4525–4548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Research Materials
Miscellaneous

