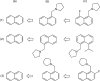
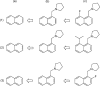
2D depiction of fragment hierarchies
- PMID: 20038186
- PMCID: PMC2810838
- DOI: 10.1021/ci900350h
2D depiction of fragment hierarchies
Abstract
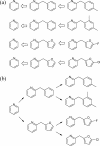
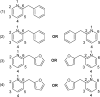
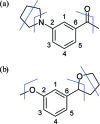
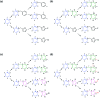
Drug discovery projects often involve organizing compounds in the form of a hierarchical tree, where each node is a substructure fragment shared by all of its descendent nodes. A method is described for producing 2D depiction layout coordinates for each of the nodes in such a tree, ensuring that common fragments within molecular structures are drawn in an identical way, and arranged with a consistent orientation. This is achieved by first deriving a common numbering scheme for common fragments, then using this scheme to redepict each of the molecules, one fragment at a time, so that common fragments have common depiction motifs. Once complete, the distinct root branches can be overlaid onto each other, after which all of the fragments and whole molecules have a common layout and orientation. Several methods are described for preparing visual representations of molecular structure hierarchies alongside activity information. Combining high level tree display and structure depiction showing common features readily facilitates insight into structure-activity relationships.
Figures





Similar articles
-
New imidazolone derivatives comprising a benzoate or sulfonamide moiety as anti-inflammatory and antibacterial inhibitors: Design, synthesis, selective COX-2, DHFR and molecular-modeling study.Bioorg Chem. 2020 Jun;99:103438. doi: 10.1016/j.bioorg.2019.103438. Epub 2019 Nov 13. Bioorg Chem. 2020. PMID: 31796216
-
Discovery of new antimalarial agents: Second-generation dual inhibitors against FP-2 and PfDHFR via fragments assembely.Bioorg Med Chem. 2017 Dec 15;25(24):6467-6478. doi: 10.1016/j.bmc.2017.10.017. Epub 2017 Oct 16. Bioorg Med Chem. 2017. PMID: 29111368
-
Rational modification of the lead molecule: Enhancement in the anticancer and dihydrofolate reductase inhibitory activity.Bioorg Med Chem Lett. 2016 Apr 15;26(8):1936-40. doi: 10.1016/j.bmcl.2016.03.015. Epub 2016 Mar 7. Bioorg Med Chem Lett. 2016. PMID: 26979156
-
DHFR Inhibitors: Reading the Past for Discovering Novel Anticancer Agents.Molecules. 2019 Mar 22;24(6):1140. doi: 10.3390/molecules24061140. Molecules. 2019. PMID: 30909399 Free PMC article. Review.
-
DNA and RNA synthesis: antifolates.Chem Rev. 2005 Feb;105(2):593-620. doi: 10.1021/cr0301144. Chem Rev. 2005. PMID: 15700958 Review. No abstract available.
Cited by
-
Basic primitives for molecular diagram sketching.J Cheminform. 2010 Oct 5;2(1):8. doi: 10.1186/1758-2946-2-8. J Cheminform. 2010. PMID: 20923555 Free PMC article.
-
Scaffold diversity of exemplified medicinal chemistry space.J Chem Inf Model. 2011 Sep 26;51(9):2174-85. doi: 10.1021/ci2001428. Epub 2011 Aug 31. J Chem Inf Model. 2011. PMID: 21877753 Free PMC article.
-
Comparative analyses of structural features and scaffold diversity for purchasable compound libraries.J Cheminform. 2017 Apr 21;9(1):25. doi: 10.1186/s13321-017-0212-4. J Cheminform. 2017. PMID: 29086044 Free PMC article.
-
Fragment virtual screening based on Bayesian categorization for discovering novel VEGFR-2 scaffolds.Mol Divers. 2015 Nov;19(4):895-913. doi: 10.1007/s11030-015-9592-4. Epub 2015 May 29. Mol Divers. 2015. PMID: 26022686
-
Machines first, humans second: on the importance of algorithmic interpretation of open chemistry data.J Cheminform. 2015 Mar 22;7:9. doi: 10.1186/s13321-015-0057-7. eCollection 2015. J Cheminform. 2015. PMID: 25798198 Free PMC article.
References
-
- Agrafiotis D. K.; Shemanarev M.; Connolly P. J.; Farnum M.; Lobanov V. S. SAR Maps: A New SAR Visualization Technique for Medicinal Chemists. J. Med. Chem. 2007, 50, 5926–5937. - PubMed
- Guha R.; Van Drie J. H. Structure−Activity Landscape Index: Identifying and Quantifying Activity Cliffs. J. Chem. Inf. Model. 2008, 48, 646–658. - PubMed
- Guha R.; Van Drie J. H. Assessing How Well a Modeling Protocol Captures a Structure−Activity Landscape. J. Chem. Inf. Model. 2008, 48, 1716–1728. - PubMed
- Lounkine E.; Auer J.; Bajorath J. Formal Concept Analysis for the Identification of Molecular Fragment Combinations Specific for Active and Highly Potent Compounds. J. Med. Chem. 2008, 51, 5342–5348. - PubMed
- Wawer M.; Peltason L.; Weskamp N.; Teckentrup A.; Bajorath J. Structure−Activity Relationship Anatomy by Network-like Similarity Graphs and Local Structure−Activity Relationship Indices. J. Med. Chem. 2008, 51, 6075–6084. - PubMed
- Kolpak J.; Connolly P. J.; Lobanov V. S.; Agrafiotis D. K. Enhanced SAR Maps: Expanding the Data Rendering Capabilities of a Popular Medicinal Chemistry Tool. J. Chem. Inf. Model. 2009, 49, 2221–2230. - PubMed
-
- Bemis G. W.; Murcko M. A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, 2887–2893. - PubMed
- Wilkens S. J.; Janes J.; Su A. I. HierS: Hierarchical Scaffold Clustering Using Topological Chemical Graphs. J. Med. Chem. 2005, 48, 3182–3193. - PubMed
- Shelat A. A.; Guy R. K. Scaffold composition and biological relevance of screening libraries. Nat. Chem. Biol. 2007, 3, 442–446. - PubMed
- Lounkine E.; Bajorath J. Core Trees and Consensus Fragment Sequences for Molecular Representation and Similarity Analysis. J. Chem. Inf. Model. 2008, 48, 1161–1166. - PubMed
-
- Schuffenhauer A.; Ertl P.; Roggo S.; Wetzel S.; Koch M. A.; Waldmann H. The Scaffold Tree—Visualization of the Scaffold Universe by Hierarchical Scaffold Classification. J. Chem. Inf. Model. 2007, 47, 47–58. - PubMed
-
- Weininger D. SMILES 1. Introduction and Encoding Rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

