Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents
- PMID: 20038498
- PMCID: PMC3525206
- DOI: 10.1186/1479-7364-4-2-107
Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents
Abstract
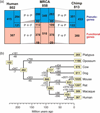
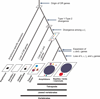
Olfaction is essential for the survival of animals. Versatile odour molecules in the environment are received by olfactory receptors (ORs), which form the largest multigene family in vertebrates. Identification of the entire repertories of OR genes using bioinformatics methods from the whole-genome sequences of diverse organisms revealed that the numbers of OR genes vary enormously, ranging from approximately 1,200 in rats and approximately 400 in humans to approximately 150 in zebrafish and approximately 15 in pufferfish. Most species have a considerable fraction of pseudogenes. Extensive phylogenetic analyses have suggested that the numbers of gene gains and losses are extremely large in the OR gene family, which is a striking example of the birth-and-death evolution. It appears that OR gene repertoires change dynamically, depending on each organism's living environment. For example, higher primates equipped with a well-developed vision system have lost a large number of OR genes. Moreover, two groups of OR genes for detecting airborne odorants greatly expanded after the time of terrestrial adaption in the tetrapod lineage, whereas fishes retain diverse repertoires of genes that were present in aquatic ancestral species. The origin of vertebrate OR genes can be traced back to the common ancestor of all chordate species, but insects, nematodes and echinoderms utilise distinctive families of chemoreceptors, suggesting that chemoreceptor genes have evolved many times independently in animal evolution.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

