Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit
- PMID: 20038530
- PMCID: PMC2820894
- DOI: 10.1128/MCB.00999-09
Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit
Abstract
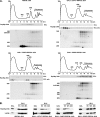
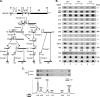
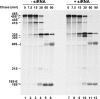
Ribosome biogenesis in eukaryotes is a major cellular activity mobilizing the products of over 200 transcriptionally coregulated genes referred to as the rRNA and ribosome biosynthesis regulon. We investigated the function of an essential, uncharacterized gene of this regulon, renamed RRP36. We show that the Rrp36p protein is nucleolar and interacts with 90S and pre-40S preribosomal particles. Its depletion affects early cleavages of the 35S pre-rRNA and results in a rapid decrease in mature 18S rRNA levels. Rrp36p is a novel component of the 90S preribosome, the assembly of which has been suggested to result from the stepwise incorporation of several modules, including the tUTP/UTP-A, PWP2/UTP-B, and UTP-C subcomplexes. We show that Rrp36p depletion does not impair the incorporation of these subcomplexes and the U3 small nucleolar RNP into preribosomes. In contrast, depletion of components of the UTP-A or UTP-B modules, but not Rrp5p, prevents Rrp36p recruitment and reduces its accumulation levels. In parallel, we studied the human orthologue of Rrp36p in HeLa cells, and we show that the function of this protein in early cleavages of the pre-rRNA has been conserved through evolution in eukaryotes.
Figures



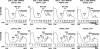

References
-
- Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
