H19 imprinting control region methylation requires an imprinted environment only in the male germ line
- PMID: 20038532
- PMCID: PMC2820884
- DOI: 10.1128/MCB.00575-09
H19 imprinting control region methylation requires an imprinted environment only in the male germ line
Abstract
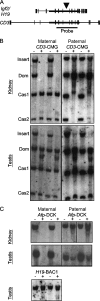
The 2.4-kb H19 imprinting control region (H19ICR) is required to establish parent-of-origin-specific epigenetic marks and expression patterns at the Igf2/H19 locus. H19ICR activity is regulated by DNA methylation. The ICR is methylated in sperm but not in oocytes, and this paternal chromosome-specific methylation is maintained throughout development. We recently showed that the H19ICR can work as an ICR even when inserted into the normally nonimprinted alpha fetoprotein locus. Paternal but not maternal copies of the ICR become methylated in somatic tissue. However, the ectopic ICR remains unmethylated in sperm. To extend these findings and investigate the mechanisms that lead to methylation of the H19ICR in the male germ line, we characterized novel mouse knock-in lines. Our data confirm that the 2.4-kb element is an autonomously acting ICR whose function is not dependent on germ line methylation. Ectopic ICRs become methylated in the male germ line, but the timing of methylation is influenced by the insertion site and by additional genetic information. Our results support the idea that DNA methylation is not the primary genomic imprint and that the H19ICR insertion is sufficient to transmit parent-of-origin-dependent DNA methylation patterns independent of its methylation status in sperm.
Figures


Similar articles
-
A randomly integrated transgenic H19 imprinting control region acquires methylation imprinting independently of its establishment in germ cells.Mol Cell Biol. 2009 Sep;29(17):4595-603. doi: 10.1128/MCB.00275-09. Epub 2009 Jun 22. Mol Cell Biol. 2009. PMID: 19546235 Free PMC article.
-
H19ICR mediated transcriptional silencing does not require target promoter methylation.Biochem Biophys Res Commun. 2016 Jul 29;476(3):121-6. doi: 10.1016/j.bbrc.2016.05.042. Epub 2016 May 10. Biochem Biophys Res Commun. 2016. PMID: 27178213 Free PMC article.
-
De novo DNA methylation through the 5'-segment of the H19 ICR maintains its imprint during early embryogenesis.Development. 2015 Nov 15;142(22):3833-44. doi: 10.1242/dev.126003. Epub 2015 Sep 28. Development. 2015. PMID: 26417043
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
-
Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation.J Biochem. 2000 May;127(5):711-5. doi: 10.1093/oxfordjournals.jbchem.a022661. J Biochem. 2000. PMID: 10788777 Review.
Cited by
-
Transgenic epigenetics: using transgenic organisms to examine epigenetic phenomena.Genet Res Int. 2012;2012:689819. doi: 10.1155/2012/689819. Epub 2012 Mar 27. Genet Res Int. 2012. PMID: 22567397 Free PMC article.
-
The chicken HS4 insulator element does not protect the H19 ICR from differential DNA methylation in yeast artificial chromosome transgenic mouse.PLoS One. 2013 Sep 4;8(9):e73925. doi: 10.1371/journal.pone.0073925. eCollection 2013. PLoS One. 2013. PMID: 24023912 Free PMC article.
-
More than insulator: multiple roles of CTCF at the H19-Igf2 imprinted domain.Front Genet. 2012 Oct 15;3:214. doi: 10.3389/fgene.2012.00214. eCollection 2012. Front Genet. 2012. PMID: 23087708 Free PMC article.
-
Fate of methylated/unmethylated H19 imprinting control region after paternal and maternal pronuclear injection.Exp Anim. 2017 Oct 30;66(4):367-378. doi: 10.1538/expanim.17-0031. Epub 2017 Jun 30. Exp Anim. 2017. PMID: 28674270 Free PMC article.
-
Long noncoding RNA H19 - a new player in the pathogenesis of liver diseases.Transl Res. 2021 Apr;230:139-150. doi: 10.1016/j.trsl.2020.11.010. Epub 2020 Nov 20. Transl Res. 2021. PMID: 33227504 Free PMC article. Review.
References
-
- Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19-gene. Genes Dev. 7:1663-1673. - PubMed
-
- Bartolomei, M. S., S. Zemel, and S. M. Tilghman. 1991. Parental imprinting of the mouse H19 gene. Nature 351:153-155. - PubMed
-
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. - PubMed
-
- Cerrato, F., A. Sparago, G. Verde, A. De Crescenzo, V. Citro, M. V. Cubellis, M. M. Rinaldi, L. Boccuto, G. Neri, C. Magnani, P. D'Angelo, P. Collini, D. Perotti, G. Sebastio, E. R. Maher, and A. Riccio. 2008. Different mechanisms cause imprinting defects at the IGF2/H19 locus in Beckwith-Wiedemann syndrome and Wilms' tumour. Hum. Mol. Genet. 17:1427-1435. - PubMed
-
- Ciccone, D. N., H. Su, S. Hevi, F. Gay, H. Lei, J. Bajko, G. L. Xu, E. Li, and T. P. Chen. 2009. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461:415-418. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
