Contribution of asparagine residues to the stabilization of a proteinaceous antigen-antibody complex, HyHEL-10-hen egg white lysozyme
- PMID: 20038580
- PMCID: PMC2844214
- DOI: 10.1074/jbc.M109.089623
Contribution of asparagine residues to the stabilization of a proteinaceous antigen-antibody complex, HyHEL-10-hen egg white lysozyme
Abstract
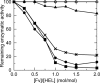
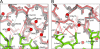
Many germ line antibodies have asparagine residues at specific sites to achieve specific antigen recognition. To study the role of asparagine residues in the stabilization of antigen-antibody complexes, we examined the interaction between hen egg white lysozyme (HEL) and the corresponding HyHEL-10 variable domain fragment (Fv). We introduced Ala and Asp substitutions into the Fv side chains of L-Asn-31, L-Asn-32, and L-Asn-92, which interact directly with residues in HEL via hydrogen bonding in the wild-type Fv-HEL complex, and we investigated the interactions between these mutant antibodies and HEL. Isothermal titration calorimetric analysis showed that all the mutations decreased the negative enthalpy change and decreased the association constants of the interaction. Structural analyses showed that the effects of the mutations on the structure of the complex could be compensated for by conformational changes and/or by gains in other interactions. Consequently, the contribution of two hydrogen bonds was minor, and their abolition by mutation resulted in only a slight decrease in the affinity of the antibody for its antigen. By comparison, the other two hydrogen bonds buried at the interfacial area had large enthalpic advantage, despite entropic loss that was perhaps due to stiffening of the interface by the bonds, and were crucial to the strength of the interaction. Deletion of these strong hydrogen bonds could not be compensated for by other structural changes. Our results suggest that asparagine can provide the two functional groups for strong hydrogen bond formation, and their contribution to the antigen-antibody interaction can be attributed to their limited flexibility and accessibility at the complex interface.
Figures


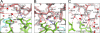
References
-
- Böhm H. J., Schneider G. (eds) (2003) Protein-Ligand Interactions: From Molecular Recognition to Drug Design (Methods and Principles in Medicinal Chemistry) Vol. 19, 1st Ed., pp. 21–51, Wiley-VCH, Weinheim, Germany
-
- Kabat E. A., Wu T. T, Bilofsky H. (1977) J. Biol. Chem. 252, 6609–6616 - PubMed
-
- Padlan E. A. (1990) Proteins Struct. Funct. Genet. 7, 112–124 - PubMed
-
- Mian I. S., Bradwell A. R., Olson A. J. (1991) J. Mol. Biol. 217, 133–151 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

