Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB
- PMID: 20038590
- PMCID: PMC2820869
- DOI: 10.1128/JB.01495-09
Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB
Abstract
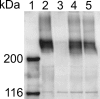
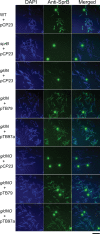
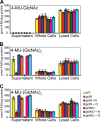
Cells of the gliding bacterium Flavobacterium johnsoniae move rapidly over surfaces. Mutations in gldN cause a partial defect in gliding. A novel bacteriophage selection strategy was used to aid construction of a strain with a deletion spanning gldN and the closely related gene gldO in an otherwise wild-type F. johnsoniae UW101 background. Bacteriophage transduction was used to move a gldN mutation into F. johnsoniae UW101 to allow phenotypic comparison with the gldNO deletion mutant. Cells of the gldN mutant formed nonspreading colonies on agar but retained some ability to glide in wet mounts. In contrast, cells of the gldNO deletion mutant were completely nonmotile, indicating that cells require GldN, or the GldN-like protein GldO, to glide. Recent results suggest that Porphyromonas gingivalis PorN, which is similar in sequence to GldN, has a role in protein secretion across the outer membrane. Cells of the F. johnsoniae gldNO deletion mutant were defective in localization of the motility protein SprB to the cell surface, suggesting that GldN may be involved in secretion of components of the motility machinery. Cells of the gldNO deletion mutant were also deficient in chitin utilization and were resistant to infection by bacteriophages, phenotypes that may also be related to defects in protein secretion.
Figures






Similar articles
-
Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system.J Bacteriol. 2015 Jan 1;197(1):147-58. doi: 10.1128/JB.02085-14. Epub 2014 Oct 20. J Bacteriol. 2015. PMID: 25331433 Free PMC article.
-
Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF.J Bacteriol. 2011 Feb;193(3):599-610. doi: 10.1128/JB.01203-10. Epub 2010 Dec 3. J Bacteriol. 2011. PMID: 21131497 Free PMC article.
-
Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis.J Bacteriol. 2005 Oct;187(20):6943-52. doi: 10.1128/JB.187.20.6943-6952.2005. J Bacteriol. 2005. PMID: 16199564 Free PMC article.
-
Cytophaga-flavobacterium gliding motility.J Mol Microbiol Biotechnol. 2004;7(1-2):63-71. doi: 10.1159/000077870. J Mol Microbiol Biotechnol. 2004. PMID: 15170404 Review.
-
Flavobacterium gliding motility and the type IX secretion system.Curr Opin Microbiol. 2015 Dec;28:72-7. doi: 10.1016/j.mib.2015.07.016. Epub 2015 Oct 23. Curr Opin Microbiol. 2015. PMID: 26461123 Review.
Cited by
-
Structural Insights into the PorK and PorN Components of the Porphyromonas gingivalis Type IX Secretion System.PLoS Pathog. 2016 Aug 10;12(8):e1005820. doi: 10.1371/journal.ppat.1005820. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27509186 Free PMC article.
-
Type IX Secretion System Effectors and Virulence of the Model Flavobacterium columnare Strain MS-FC-4.Appl Environ Microbiol. 2022 Feb 8;88(3):e0170521. doi: 10.1128/AEM.01705-21. Epub 2021 Nov 24. Appl Environ Microbiol. 2022. PMID: 34818105 Free PMC article.
-
The Type 9 Secretion System Is Required for Flavobacterium johnsoniae Biofilm Formation.Front Microbiol. 2021 Sep 2;12:660887. doi: 10.3389/fmicb.2021.660887. eCollection 2021. Front Microbiol. 2021. PMID: 34539591 Free PMC article.
-
The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function.Front Cell Infect Microbiol. 2017 May 26;7:215. doi: 10.3389/fcimb.2017.00215. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28603700 Free PMC article. Review.
-
Diverse C-Terminal Sequences Involved in Flavobacterium johnsoniae Protein Secretion.J Bacteriol. 2017 May 25;199(12):e00884-16. doi: 10.1128/JB.00884-16. Print 2017 Jun 15. J Bacteriol. 2017. PMID: 28396348 Free PMC article.
References
-
- Duchaud, E., M. Boussaha, V. Loux, J. F. Bernardet, C. Michel, B. Kerouault, S. Mondot, P. Nicolas, R. Bossy, C. Caron, P. Bessières, J. F. Gibrat, S. Claverol, F. Dumetz, M. L. Hénaff, and A. Benmansour. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat. Biotechnol. 25:763-769. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

