Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes
- PMID: 20038606
- PMCID: PMC2837988
- DOI: 10.1128/EC.00348-09
Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes
Abstract
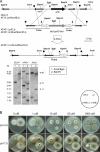
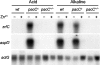
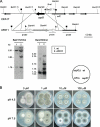
Aspergillus fumigatus has three zinc transporter-encoding genes whose expression is regulated by both pH and the environmental concentration of zinc. We have previously reported that the zrfA and zrfB genes of A. fumigatus are transcribed at higher levels and are required for fungal growth under acidic zinc-limiting conditions whereas they are dispensable for growth in neutral or alkaline zinc-limiting media. Here we report that the transporter of the zinc uptake system that functions in A. fumigatus growing in neutral or alkaline environments is encoded by zrfC. The transcription of zrfC occurs divergently with respect to the adjacent aspf2 gene, which encodes an immunodominant antigen secreted by A. fumigatus. The two genes-zrfC and aspf2-are required to different extents for fungal growth in alkaline and extreme zinc-limiting media. Indeed, these environmental conditions induce the simultaneous transcription of both genes mediated by the transcriptional regulators ZafA and PacC. ZafA upregulates the expression of zrfC and aspf2 under zinc-limiting conditions regardless of the ambient pH, whereas PacC represses the expression of these genes under acidic growth conditions. Interestingly, the mode of action of PacC for zrfC-aspf2 transcription contrasts with the more widely accepted model for PacC function, according to which under alkaline growth conditions PacC would activate the transcription of alkaline-expressed genes but would repress the transcription of acid-expressed genes. In sum, this report provides a good framework for investigating several important aspects of the biology of species of Aspergillus, including the repression of alkaline genes by PacC at acidic pH and the interrelationship that must exist between tissue pH, metal availability in the host tissue, and fungal virulence.
Figures






References
-
- Amich J., Leal F., Calera J. A. 2009. Repression of the acid ZrfA/ZrfB zinc-uptake system of Aspergillus fumigatus mediated by PacC under neutral, zinc-limiting conditions. Int. Microbiol. 12:39–47 - PubMed
-
- Auld D. S. 2001. Zinc coordination sphere in biochemical zinc sites. Biometals 14:271–313 - PubMed
-
- Basta N. T., Ryan J. A., Chaney R. L. 2005. Trace element chemistry in residual-treated soil: key concepts and metal bioavailability. J. Environ. Qual. 34:49–63 - PubMed
-
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

