Comparison of methods for profiling O-glycosylation: Human Proteome Organisation Human Disease Glycomics/Proteome Initiative multi-institutional study of IgA1
- PMID: 20038609
- PMCID: PMC2860227
- DOI: 10.1074/mcp.M900450-MCP200
Comparison of methods for profiling O-glycosylation: Human Proteome Organisation Human Disease Glycomics/Proteome Initiative multi-institutional study of IgA1
Abstract
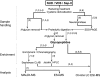
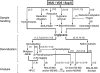
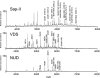
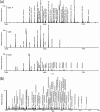
The Human Proteome Organisation Human Disease Glycomics/Proteome Initiative recently coordinated a multi-institutional study that evaluated methodologies that are widely used for defining the N-glycan content in glycoproteins. The study convincingly endorsed mass spectrometry as the technique of choice for glycomic profiling in the discovery phase of diagnostic research. The present study reports the extension of the Human Disease Glycomics/Proteome Initiative's activities to an assessment of the methodologies currently used for O-glycan analysis. Three samples of IgA1 isolated from the serum of patients with multiple myeloma were distributed to 15 laboratories worldwide for O-glycomics analysis. A variety of mass spectrometric and chromatographic procedures representative of current methodologies were used. Similar to the previous N-glycan study, the results convincingly confirmed the pre-eminent performance of MS for O-glycan profiling. Two general strategies were found to give the most reliable data, namely direct MS analysis of mixtures of permethylated reduced glycans in the positive ion mode and analysis of native reduced glycans in the negative ion mode using LC-MS approaches. In addition, mass spectrometric methodologies to analyze O-glycopeptides were also successful.
Figures

References
-
- Wada Y., Azadi P., Costello C. E., Dell A., Dwek R. A., Geyer H., Geyer R., Kakehi K., Karlsson N. G., Kato K., Kawasaki N., Khoo K. H., Kim S., Kondo A., Lattova E., Mechref Y., Miyoshi E., Nakamura K., Narimatsu H., Novotny M. V., Packer N. H., Perreault H., Peter-Katalinic J., Pohlentz G., Reinhold V. N., Rudd P. M., Suzuki A., Taniguchi N. (2007) Comparison of the methods for profiling glycoprotein glycans—HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology 17, 411–422 - PubMed
-
- Mestecky J. (1988) Immunobiology of IgA. Am. J. Kidney Dis 12, 378–383 - PubMed
-
- Baenziger J., Kornfeld S. (1974) Structure of the carbohydrate units of IgA1 immunoglobulin. II. Structure of the O-glycosidically linked oligosaccharide units. J. Biol. Chem 249, 7270–7281 - PubMed
-
- Field M. C., Dwek R. A., Edge C. J., Rademacher T. W. (1989) O-linked oligosaccharides from human serum immunoglobulin A1. Biochem. Soc. Trans 17, 1034–1035 - PubMed
-
- Mattu T. S., Pleass R. J., Willis A. C., Kilian M., Wormald M. R., Lellouch A. C., Rudd P. M., Woof J. M., Dwek R. A. (1998) The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fc alpha receptor interactions. J. Biol. Chem 273, 2260–2272 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK082753/DK/NIDDK NIH HHS/United States
- P41 RR10888/RR/NCRR NIH HHS/United States
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- S10 RR020946/RR/NCRR NIH HHS/United States
- R01 DK078244/DK/NIDDK NIH HHS/United States
- R01 DK071802/DK/NIDDK NIH HHS/United States
- DK078244/DK/NIDDK NIH HHS/United States
- P41 RR010888/RR/NCRR NIH HHS/United States
- DK80301/DK/NIDDK NIH HHS/United States
- R01 GM098539/GM/NIGMS NIH HHS/United States
- S10 RR20946/RR/NCRR NIH HHS/United States
- DK71802/DK/NIDDK NIH HHS/United States
- DK077279/DK/NIDDK NIH HHS/United States
- DK064400/DK/NIDDK NIH HHS/United States
- R24 DK064400/DK/NIDDK NIH HHS/United States
- R21 DK080301/DK/NIDDK NIH HHS/United States
- S10 RR15942/RR/NCRR NIH HHS/United States
- R01 DK082753/DK/NIDDK NIH HHS/United States
- R56 DK078244/DK/NIDDK NIH HHS/United States
- R21 DK077279/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

