Genes involved in long-chain alkene biosynthesis in Micrococcus luteus
- PMID: 20038703
- PMCID: PMC2820947
- DOI: 10.1128/AEM.02312-09
Genes involved in long-chain alkene biosynthesis in Micrococcus luteus
Abstract
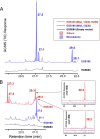
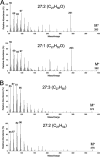
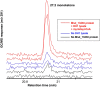
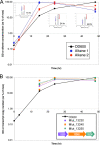
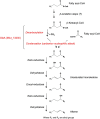
Aliphatic hydrocarbons are highly appealing targets for advanced cellulosic biofuels, as they are already predominant components of petroleum-based gasoline and diesel fuels. We have studied alkene biosynthesis in Micrococcus luteus ATCC 4698, a close relative of Sarcina lutea (now Kocuria rhizophila), which 4 decades ago was reported to biosynthesize iso- and anteiso-branched, long-chain alkenes. The underlying biochemistry and genetics of alkene biosynthesis were not elucidated in those studies. We show here that heterologous expression of a three-gene cluster from M. luteus (Mlut_13230-13250) in a fatty acid-overproducing Escherichia coli strain resulted in production of long-chain alkenes, predominantly 27:3 and 29:3 (no. carbon atoms: no. C=C bonds). Heterologous expression of Mlut_13230 (oleA) alone produced no long-chain alkenes but unsaturated aliphatic monoketones, predominantly 27:2, and in vitro studies with the purified Mlut_13230 protein and tetradecanoyl-coenzyme A (CoA) produced the same C(27) monoketone. Gas chromatography-time of flight mass spectrometry confirmed the elemental composition of all detected long-chain alkenes and monoketones (putative intermediates of alkene biosynthesis). Negative controls demonstrated that the M. luteus genes were responsible for production of these metabolites. Studies with wild-type M. luteus showed that the transcript copy number of Mlut_13230-13250 and the concentrations of 29:1 alkene isomers (the dominant alkenes produced by this strain) generally corresponded with bacterial population over time. We propose a metabolic pathway for alkene biosynthesis starting with acyl-CoA (or-ACP [acyl carrier protein]) thioesters and involving decarboxylative Claisen condensation as a key step, which we believe is catalyzed by OleA. Such activity is consistent with our data and with the homology (including the conserved Cys-His-Asn catalytic triad) of Mlut_13230 (OleA) to FabH (beta-ketoacyl-ACP synthase III), which catalyzes decarboxylative Claisen condensation during fatty acid biosynthesis.
Figures

References
-
- Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain, nonisoprenoid hydrocarbons. I. Characterization of the hydrocarbons of Sarcina lutea and the isolation of possible intermediates of biosynthesis. Biochemistry 8:394-404. - PubMed
-
- Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain, nonisoprenoid hydrocarbons. IV. Characteristics of synthesis by a cell-free preparation of Sarcina lutea. Biochemistry 8:3317-3324. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

