DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes
- PMID: 20038729
- PMCID: PMC2815995
- DOI: 10.1200/JCO.2009.23.4781
DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes
Erratum in
- J Clin Oncol. 2010 Jun 20;28(18):3098
Abstract
Purpose: The current classification systems of myelodysplastic syndromes (MDS), including the International Prognostic Scoring System (IPSS), do not fully reflect the molecular heterogeneity of the disease. Molecular characterization may predict clinical outcome and help stratify patients for targeted therapies. Epigenetic therapy using decitabine, a DNA hypomethylating agent, is clinically effective for the treatment of MDS. Therefore, we investigated the association between DNA methylation and clinical outcome in MDS.
Patients and methods: We screened 24 patients with MDS for promoter CpG island methylation of 24 genes and identified aberrant hypermethylation at 10 genes. We then performed quantitative methylation analyses by bisulfite pyrosequencing of the identified genes in 317 patient samples from three independent studies and assessed relations between methylation and clinical outcome.
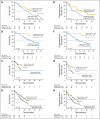
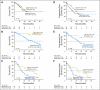
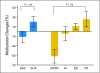
Results: In an initial training cohort of 89 patients with MDS, methylation frequencies of individual genes ranged from 7% to 70% and were highly concordant. Therefore, we defined a methylation z score based on all genes for each patient. We found that patients with higher levels of methylation, compared with patients with lower levels, had a shorter median overall survival (12.3 v 17.5 months, respectively; P = .04) and shorter median progression-free survival (6.4 v 14.9 months, respectively; P = .009). This methylation prognostic model was independent of age, sex, and IPSS group. Applied to two validation cohorts (228 patients), this model was confirmed as an independent prognostic predictor for survival. Although methylation at baseline did not correlate with clinical response to decitabine, we observed a significant correlation between reduced methylation over time and clinical responses.
Conclusion: DNA methylation predicts overall and progression-free survival in MDS.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures
Similar articles
-
Pyrosequencing quantified methylation level of miR-124 predicts shorter survival for patients with myelodysplastic syndrome.Clin Epigenetics. 2017 Aug 30;9:91. doi: 10.1186/s13148-017-0388-5. eCollection 2017. Clin Epigenetics. 2017. PMID: 28861128 Free PMC article.
-
Promoter hypermethylation of p15INK4B, HIC1, CDH1, and ER is frequent in myelodysplastic syndrome and predicts poor prognosis in early-stage patients.Eur J Haematol. 2006 Jan;76(1):23-32. doi: 10.1111/j.1600-0609.2005.00559.x. Eur J Haematol. 2006. PMID: 16343268
-
Prognostic role of immunohistochemical analysis of 5 mc in myelodysplastic syndromes.Eur J Haematol. 2013 Sep;91(3):219-227. doi: 10.1111/ejh.12145. Epub 2013 Jun 28. Eur J Haematol. 2013. PMID: 23679560
-
DNA methylation and demethylating drugs in myelodysplastic syndromes and secondary leukemias.Haematologica. 2002 Dec;87(12):1324-41. Haematologica. 2002. PMID: 12495905 Review.
-
Clinical results with the DNA hypomethylating agent 5-aza-2'-deoxycytidine (decitabine) in patients with myelodysplastic syndromes: an update.Semin Hematol. 2012 Oct;49(4):330-41. doi: 10.1053/j.seminhematol.2012.08.001. Semin Hematol. 2012. PMID: 23079063 Review.
Cited by
-
Specific molecular signatures predict decitabine response in chronic myelomonocytic leukemia.J Clin Invest. 2015 May;125(5):1857-72. doi: 10.1172/JCI78752. Epub 2015 Mar 30. J Clin Invest. 2015. PMID: 25822018 Free PMC article. Clinical Trial.
-
Mutations in epigenetic regulators in myelodysplastic syndromes.Int J Hematol. 2012 Jan;95(1):8-16. doi: 10.1007/s12185-011-0996-3. Epub 2012 Jan 11. Int J Hematol. 2012. PMID: 22234528 Review.
-
Factors affecting the persistence of drug-induced reprogramming of the cancer methylome.Epigenetics. 2016 Apr 2;11(4):273-87. doi: 10.1080/15592294.2016.1158364. Epub 2016 Apr 15. Epigenetics. 2016. PMID: 27082926 Free PMC article.
-
Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial.Lancet Oncol. 2020 Nov;21(11):1433-1442. doi: 10.1016/S1470-2045(20)30441-1. Epub 2020 Oct 6. Lancet Oncol. 2020. PMID: 33035457 Free PMC article. Clinical Trial.
-
DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia.Epigenetics. 2014 Apr;9(4):503-12. doi: 10.4161/epi.27644. Epub 2014 Jan 17. Epigenetics. 2014. PMID: 24441198 Free PMC article.
References
-
- Gilliland DG. Hematologic malignancies. Curr Opin Hematol. 2001;8:189–191. - PubMed
-
- Komrokji RS, Bennett JM. Evolving classifications of the myelodysplastic syndromes. Curr Opin Hematol. 2007;14:98–105. - PubMed
-
- Valent P, Horny HP, Bennett JM, et al. Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: Consensus statements and report from a working conference. Leuk Res. 2007;31:727–736. - PubMed
-
- Haase D, Germing U, Schanz J, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: Evidence from a core dataset of 2124 patients. Blood. 2007;110:4385–4395. - PubMed
-
- Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer. 2006;106:1794–1803. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

