The evolutionarily conserved TSC/Rheb pathway activates Notch in tuberous sclerosis complex and Drosophila external sensory organ development
- PMID: 20038815
- PMCID: PMC2798691
- DOI: 10.1172/JCI40221
The evolutionarily conserved TSC/Rheb pathway activates Notch in tuberous sclerosis complex and Drosophila external sensory organ development
Abstract
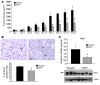
Mutations in either of the genes encoding the tuberous sclerosis complex (TSC), TSC1 and TSC2, result in a multisystem tumor disorder characterized by lesions with unusual lineage expression patterns. How these unusual cell-fate determination patterns are generated is unclear. We therefore investigated the role of the TSC in the Drosophila external sensory organ (ESO), a classic model of asymmetric cell division. In normal development, the sensory organ precursor cell divides asymmetrically through differential regulation of Notch signaling to produce a pIIa and a pIIb cell. We report here that inactivation of Tsc1 and overexpression of the Ras homolog Rheb each resulted in duplication of the bristle and socket cells, progeny of the pIIa cell, and loss of the neuronal cell, a product of pIIb cell division. Live imaging of ESO development revealed this cell-fate switch occurred at the pIIa-pIIb 2-cell stage. In human angiomyolipomas, benign renal neoplasms often found in tuberous sclerosis patients, we found evidence of Notch receptor cleavage and Notch target gene activation. Further, an angiomyolipoma-derived cell line carrying biallelic TSC2 mutations exhibited TSC2- and Rheb-dependent Notch activation. Finally, inhibition of Notch signaling using a gamma-secretase inhibitor suppressed proliferation of Tsc2-null rat cells in a xenograft model. Together, these data indicate that the TSC and Rheb regulate Notch-dependent cell-fate decision in Drosophila and Notch activity in mammalian cells and that Notch dysregulation may underlie some of the distinctive clinical and pathologic features of TSC.
Figures
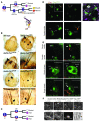
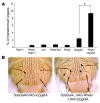
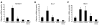


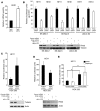
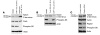
Comment in
-
New roles for Notch in tuberous sclerosis.J Clin Invest. 2010 Jan;120(1):84-7. doi: 10.1172/JCI41897. Epub 2009 Dec 28. J Clin Invest. 2010. PMID: 20038806 Free PMC article.
Similar articles
-
New roles for Notch in tuberous sclerosis.J Clin Invest. 2010 Jan;120(1):84-7. doi: 10.1172/JCI41897. Epub 2009 Dec 28. J Clin Invest. 2010. PMID: 20038806 Free PMC article.
-
Rhebbing up mTOR: new insights on TSC1 and TSC2, and the pathogenesis of tuberous sclerosis.Cancer Biol Ther. 2003 Sep-Oct;2(5):471-6. doi: 10.4161/cbt.2.5.446. Cancer Biol Ther. 2003. PMID: 14614311 Review.
-
Frequent [corrected] hyperphosphorylation of ribosomal protein S6 [corrected] in lymphangioleiomyomatosis-associated angiomyolipomas.Mod Pathol. 2006 Jun;19(6):839-46. doi: 10.1038/modpathol.3800610. Mod Pathol. 2006. PMID: 16575396
-
TSC1/2 regulates intestinal stem cell maintenance and lineage differentiation through Rheb-TORC1-S6K but independently of nutritional status or Notch regulation.J Cell Sci. 2013 Sep 1;126(Pt 17):3884-92. doi: 10.1242/jcs.125294. Epub 2013 Jul 10. J Cell Sci. 2013. PMID: 23843608
-
The mTOR/S6K signalling pathway: the role of the TSC1/2 tumour suppressor complex and the proto-oncogene Rheb.Novartis Found Symp. 2004;262:148-54; discussion 154-9, 265-8. Novartis Found Symp. 2004. PMID: 15562827 Review.
Cited by
-
Cooperative benefit for the combination of rapamycin and imatinib in tuberous sclerosis complex neoplasia.Vasc Cell. 2012 Jul 5;4(1):11. doi: 10.1186/2045-824X-4-11. Vasc Cell. 2012. PMID: 22765013 Free PMC article.
-
A circuitry and biochemical basis for tuberous sclerosis symptoms: from epilepsy to neurocognitive deficits.Int J Dev Neurosci. 2013 Nov;31(7):667-78. doi: 10.1016/j.ijdevneu.2013.02.008. Epub 2013 Feb 26. Int J Dev Neurosci. 2013. PMID: 23485365 Free PMC article. Review.
-
Are oncogenes sufficient to cause human cancer?Proc Natl Acad Sci U S A. 2010 Nov 30;107(48):20599-600. doi: 10.1073/pnas.1015563107. Epub 2010 Nov 17. Proc Natl Acad Sci U S A. 2010. PMID: 21084639 Free PMC article. No abstract available.
-
Inhibition of the mechanistic target of rapamycin induces cell survival via MAPK in tuberous sclerosis complex.Orphanet J Rare Dis. 2020 Aug 17;15(1):209. doi: 10.1186/s13023-020-01490-w. Orphanet J Rare Dis. 2020. PMID: 32807195 Free PMC article.
-
High-throughput drug screen identifies chelerythrine as a selective inducer of death in a TSC2-null setting.Mol Cancer Res. 2015 Jan;13(1):50-62. doi: 10.1158/1541-7786.MCR-14-0440. Epub 2014 Sep 3. Mol Cancer Res. 2015. PMID: 25185584 Free PMC article.
References
-
- Crino PB, Henske EP. New developments in the neurobiology of the tuberous sclerosis complex. Neurology. 1999;53(7):1384–1390. - PubMed
-
- Plank TL, Yeung RS, Henske EP. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 1998;58(21):4766–4770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

