DNA interstrand crosslink repair in mammalian cells: step by step
- PMID: 20039786
- PMCID: PMC2824768
- DOI: 10.3109/10409230903501819
DNA interstrand crosslink repair in mammalian cells: step by step
Abstract
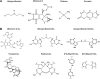
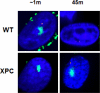
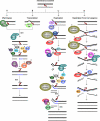
Interstrand DNA crosslinks (ICLs) are formed by natural products of metabolism and by chemotherapeutic reagents. Work in E. coli identified a two cycle repair scheme involving incisions on one strand on either side of the ICL (unhooking) producing a gapped intermediate with the incised oligonucleotide attached to the intact strand. The gap is filled by recombinational repair or lesion bypass synthesis. The remaining monoadduct is then removed by nucleotide excision repair (NER). Despite considerable effort, our understanding of each step in mammalian cells is still quite limited. In part this reflects the variety of crosslinking compounds, each with distinct structural features, used by different investigators. Also, multiple repair pathways are involved, variably operative during the cell cycle. G(1) phase repair requires functions from NER, although the mechanism of recognition has not been determined. Repair can be initiated by encounters with the transcriptional apparatus, or a replication fork. In the case of the latter, the reconstruction of a replication fork, stalled or broken by collision with an ICL, adds to the complexity of the repair process. The enzymology of unhooking, the identity of the lesion bypass polymerases required to fill the first repair gap, and the functions involved in the second repair cycle are all subjects of active inquiry. Here we will review current understanding of each step in ICL repair in mammalian cells.
Figures

References
-
- Akagi J, Masutani C, Kataoka Y, Kan T, Ohashi E, Mori T, Ohmori H, Hanaoka F. Interaction with DNA polymerase eta is required for nuclear accumulation of REV1 and suppression of spontaneous mutations in human cells. DNA Repair (Amst) 2009;8:585–599. - PubMed
-
- Allan JM, Engelward BP, Dreslin AJ, Wyatt MD, Tomasz M, Samson LD. Mammalian 3-methyladenine DNA glycosylase protects against the toxicity and clastogenicity of certain chemotherapeutic DNA cross-linking agents. Cancer Res. 1998;58:3965–3973. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
