Gene profiling of maternal hepatic adaptations to pregnancy
- PMID: 20040050
- PMCID: PMC4356012
- DOI: 10.1111/j.1478-3231.2009.02183.x
Gene profiling of maternal hepatic adaptations to pregnancy
Abstract
Background: Maternal metabolic demands change dramatically during the course of gestation and must be co-ordinated with the needs of the developing placenta and fetus. The liver is critically involved in metabolism and other important functions. However, maternal hepatic adjustments to pregnancy are poorly understood.
Aim: The aim of the study was to evaluate the influences of pregnancy on the maternal liver growth and gene expression profile.
Methods: Holtzman Sprague-Dawley rats were mated and sacrificed at various stages of gestation and post-partum. The maternal livers were analysed in gravimetric response, DNA content by PicoGreen dsDNA quantitation reagent, hepatocyte ploidy by flow cytometry and hepatocyte proliferation by ki-67 immunostaining. Gene expression profiling of non-pregnant and gestation d18.5 maternal hepatic tissue was analysed using a DNA microarray approach and partially verified by northern blot or quantitative real-time PCR analysis.
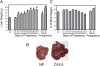
Results: During pregnancy, the liver exhibited approximately an 80% increase in size, proportional to the increase in body weight of the pregnant animals. The pregnancy-induced hepatomegaly was a physiological event of liver growth manifested by increases in maternal hepatic DNA content and hepatocyte proliferation. Pregnancy did not affect hepatocyte polyploidization. Pregnancy-dependent changes in hepatic expression were noted for a number of genes, including those associated with cell proliferation, cytokine signalling, liver regeneration and metabolism.
Conclusions: The metabolic demands of pregnancy cause marked adjustments in maternal liver physiology. Central to these adjustments are an expansion in hepatic capacity and changes in hepatic gene expression. Our findings provide insights into pregnancy-dependent hepatic adaptations.
Figures






Similar articles
-
Activation of Proneuronal Transcription Factor Ascl1 in Maternal Liver Ensures a Healthy Pregnancy.Cell Mol Gastroenterol Hepatol. 2022;13(1):35-55. doi: 10.1016/j.jcmgh.2021.08.009. Epub 2021 Aug 23. Cell Mol Gastroenterol Hepatol. 2022. PMID: 34438112 Free PMC article.
-
Effects of maternal starvation on hepatocyte proliferation in the late gestation fetal rat.Pediatr Res. 2005 Feb;57(2):185-91. doi: 10.1203/01.PDR.0000151646.55587.0F. Epub 2004 Dec 20. Pediatr Res. 2005. PMID: 15611345
-
Pregnancy and lactation modulate maternal splenic growth and development of the erythroid lineage in the rat and mouse.Reprod Fertil Dev. 2008;20(2):303-10. doi: 10.1071/rd07106. Reprod Fertil Dev. 2008. PMID: 18255020
-
Maternal creatine homeostasis is altered during gestation in the spiny mouse: is this a metabolic adaptation to pregnancy?BMC Pregnancy Childbirth. 2015 Apr 14;15:92. doi: 10.1186/s12884-015-0524-1. BMC Pregnancy Childbirth. 2015. PMID: 25885219 Free PMC article.
-
Cooperation of liver cells in health and disease.Adv Anat Embryol Cell Biol. 2001;161:III-XIII, 1-151. doi: 10.1007/978-3-642-56553-3. Adv Anat Embryol Cell Biol. 2001. PMID: 11729749 Review.
Cited by
-
Nano-titanium dioxide inhalation exposure during gestation drives redox dysregulation and vascular dysfunction across generations.Part Fibre Toxicol. 2022 Mar 9;19(1):18. doi: 10.1186/s12989-022-00457-y. Part Fibre Toxicol. 2022. PMID: 35260159 Free PMC article.
-
Activation of Proneuronal Transcription Factor Ascl1 in Maternal Liver Ensures a Healthy Pregnancy.Cell Mol Gastroenterol Hepatol. 2022;13(1):35-55. doi: 10.1016/j.jcmgh.2021.08.009. Epub 2021 Aug 23. Cell Mol Gastroenterol Hepatol. 2022. PMID: 34438112 Free PMC article.
-
Expression of melatonin receptors and CD4 in the ovine thymus, lymph node, spleen and liver during early pregnancy.Immunology. 2020 May;160(1):52-63. doi: 10.1111/imm.13180. Epub 2020 Mar 17. Immunology. 2020. PMID: 32052861 Free PMC article.
-
Genetic variants associated with ALT elevation from therapeutic acetaminophen.Clin Toxicol (Phila). 2022 Nov;60(11):1198-1204. doi: 10.1080/15563650.2022.2117053. Epub 2022 Sep 14. Clin Toxicol (Phila). 2022. PMID: 36102175 Free PMC article.
-
The Role of Placental Hormones in Mediating Maternal Adaptations to Support Pregnancy and Lactation.Front Physiol. 2018 Aug 17;9:1091. doi: 10.3389/fphys.2018.01091. eCollection 2018. Front Physiol. 2018. PMID: 30174608 Free PMC article. Review.
References
-
- Nielsen JH, Svensson C, Galsgaard ED, Moldrup A, Billestrup N. Beta cell proliferation and growth factors. J Mol Med. 1999;77(1):62–6. - PubMed
-
- Bustamante JJ, Dai G, Soares MJ. Pregnancy and lactation modulate maternal splenic growth and development of the erythroid lineage in the rat and mouse. Reproduction, fertility, and development. 2008;20(2):303–10. - PubMed
-
- Shingo T, Gregg C, Enwere E, Fujikawa H, Hassam R, Geary C, et al. Pregnancy-stimulated neurogenesis in the adult female forebrain mediated by prolactin. Science. 2003;299(5603):117–20. - PubMed
-
- Audus KL, Soares MJ, Hunt JS. Characteristics of the fetal/maternal interface with potential usefulness in the development of future immunological and pharmacological strategies. J Pharmacol Exp Ther. 2002;301(2):402–9. - PubMed
-
- Rahman TM, Wendon J. Severe hepatic dysfunction in pregnancy. Qjm. 2002;95(6):343–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

