Comprehensive in silico prediction and analysis of chlamydial outer membrane proteins reflects evolution and life style of the Chlamydiae
- PMID: 20040079
- PMCID: PMC2811131
- DOI: 10.1186/1471-2164-10-634
Comprehensive in silico prediction and analysis of chlamydial outer membrane proteins reflects evolution and life style of the Chlamydiae
Abstract
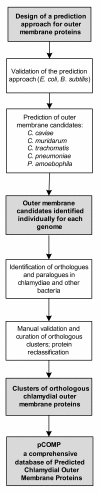
Background: Chlamydiae are obligate intracellular bacteria comprising some of the most important bacterial pathogens of animals and humans. Although chlamydial outer membrane proteins play a key role for attachment to and entry into host cells, only few have been described so far. We developed a comprehensive, multiphasic in silico approach, including the calculation of clusters of orthologues, to predict outer membrane proteins using conservative criteria. We tested this approach using Escherichia coli (positive control) and Bacillus subtilis (negative control), and applied it to five chlamydial species; Chlamydia trachomatis, Chlamydia muridarum, Chlamydia (a.k.a. Chlamydophila) pneumoniae, Chlamydia (a.k.a. Chlamydophila) caviae, and Protochlamydia amoebophila.
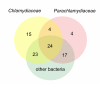
Results: In total, 312 chlamydial outer membrane proteins and lipoproteins in 88 orthologous clusters were identified, including 238 proteins not previously recognized to be located in the outer membrane. Analysis of their taxonomic distribution revealed an evolutionary conservation among Chlamydiae, Verrucomicrobia, Lentisphaerae and Planctomycetes as well as lifestyle-dependent conservation of the chlamydial outer membrane protein composition.
Conclusion: This analysis suggested a correlation between the outer membrane protein composition and the host range of chlamydiae and revealed a common set of outer membrane proteins shared by these intracellular bacteria. The collection of predicted chlamydial outer membrane proteins is available at the online database pCOMP http://www.microbial-ecology.net/pcomp and might provide future guidance in the quest for anti-chlamydial vaccines.
Figures
Similar articles
-
A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane.Cell Microbiol. 2000 Feb;2(1):35-47. doi: 10.1046/j.1462-5822.2000.00029.x. Cell Microbiol. 2000. PMID: 11207561
-
Chlamydia outer membrane protein discovery using genomics.Curr Opin Microbiol. 2001 Feb;4(1):16-20. doi: 10.1016/s1369-5274(00)00158-2. Curr Opin Microbiol. 2001. PMID: 11173028 Review.
-
Conserved features and major differences in the outer membrane protein composition of chlamydiae.Environ Microbiol. 2015 Apr;17(4):1397-413. doi: 10.1111/1462-2920.12621. Epub 2014 Oct 22. Environ Microbiol. 2015. PMID: 25212454
-
Proteomic analysis of the outer membrane of Protochlamydia amoebophila elementary bodies.Proteomics. 2010 Dec;10(24):4363-76. doi: 10.1002/pmic.201000302. Epub 2010 Nov 23. Proteomics. 2010. PMID: 21136591
-
Chlamydial polymorphic membrane proteins: regulation, function and potential vaccine candidates.Virulence. 2016;7(1):11-22. doi: 10.1080/21505594.2015.1111509. Epub 2015 Nov 18. Virulence. 2016. PMID: 26580416 Free PMC article. Review.
Cited by
-
Chlamydia pan-genomic analysis reveals balance between host adaptation and selective pressure to genome reduction.BMC Genomics. 2019 Sep 12;20(1):710. doi: 10.1186/s12864-019-6059-5. BMC Genomics. 2019. PMID: 31510914 Free PMC article.
-
Transitional forms between the three domains of life and evolutionary implications.Proc Biol Sci. 2011 Nov 22;278(1723):3321-8. doi: 10.1098/rspb.2011.1581. Epub 2011 Sep 14. Proc Biol Sci. 2011. PMID: 21920985 Free PMC article.
-
Amoebal endosymbiont Neochlamydia genome sequence illuminates the bacterial role in the defense of the host amoebae against Legionella pneumophila.PLoS One. 2014 Apr 18;9(4):e95166. doi: 10.1371/journal.pone.0095166. eCollection 2014. PLoS One. 2014. PMID: 24747986 Free PMC article.
-
Systems biology approaches to new vaccine development.Curr Opin Immunol. 2011 Jun;23(3):436-43. doi: 10.1016/j.coi.2011.04.005. Epub 2011 May 11. Curr Opin Immunol. 2011. PMID: 21570272 Free PMC article. Review.
-
Architecture and host interface of environmental chlamydiae revealed by electron cryotomography.Environ Microbiol. 2014 Feb;16(2):417-29. doi: 10.1111/1462-2920.12299. Epub 2013 Nov 8. Environ Microbiol. 2014. PMID: 24118768 Free PMC article.
References
-
- WHO. Priority eye diseases. 2008. http://www.who.int/blindness/causes/priority/en/index2.html
-
- WHO. Global prevalence and incidence of curable STIs. Geneva: World Health Organization; 2001.
-
- Mahony JB, Coombes BK, Chernesky MA. In: Manual of clinical microbiology. 8. Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editor. Vol. 1. Washington, DC: ASM Press; 2003. Chlamydia and Chlamydophila; pp. 991–1004.
-
- Kuo C-C, Horn M, Stephens RS. In: Bergey's Manual of Systematic Bacteriology - The Planctomycetes, Spriochaetes, Fibrobacteres, Bacteriodetes and Fusobacteria. 2. Hedlund B, Krieg NR, Ludwig W, Paster BJ, Staley JT, Ward N, Whitman WB, editor. New York: Springer; 2008. The order Chlamydiales. in press .
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

