HIV-1 subtype and viral tropism determination for evaluating antiretroviral therapy options: an analysis of archived Kenyan blood samples
- PMID: 20040114
- PMCID: PMC2804586
- DOI: 10.1186/1471-2334-9-215
HIV-1 subtype and viral tropism determination for evaluating antiretroviral therapy options: an analysis of archived Kenyan blood samples
Abstract
Background: Infection with HIV-1 is characterized by genetic diversity such that specific viral subtypes are predominant in specific geographical areas. The genetic variation in HIV-1 pol and env genes is responsible for rapid development of resistance to current drugs. This variation has influenced disease progression among the infected and necessitated the search for alternative drugs with novel targets. Though successfully used in developed countries, these novel drugs are still limited in resource-poor countries. The aim of this study was to determine HIV-1 subtypes, recombination, dual infections and viral tropism of HIV-1 among Kenyan patients prior to widespread use of antiretroviral drugs.
Methods: Remnant blood samples from consenting sexually transmitted infection (STI) patients in Nairobi were collected between February and May 2001 and stored. Polymerase chain reaction and cloning of portions of HIV-1 gag, pol and env genes was carried out followed by automated DNA sequencing.
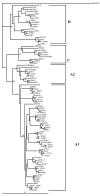
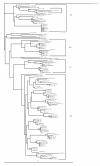
Results: Twenty HIV-1 positive samples (from 11 females and 9 males) were analyzed. The average age of males (32.5 years) and females (26.5 years) was significantly different (p value < 0.0001). Phylogenetic analysis revealed that 90% (18/20) were concordant HIV-1 subtypes: 12 were subtype A1; 2, A2; 3, D and 1, C. Two samples (10%) were discordant showing different subtypes in the three regions. Of 19 samples checked for co-receptor usage, 14 (73.7%) were chemokine co-receptor 5 (CCR5) variants while three (15.8%) were CXCR4 variants. Two had dual/mixed co-receptor use with X4 variants being minor population.
Conclusion: HIV-1 subtype A accounted for majority of the infections. Though perceived to be a high risk population, the prevalence of recombination in this sample was low with no dual infections detected. Genotypic co-receptor analysis showed that most patients harbored viruses that are predicted to use CCR5.
Figures

References
-
- Piot P, Bartos M. In: AIDS in Africa. 2. Essex M, Mboup S, Kanki PJ, Marlink PJ, Tlou SD, editor. New York. Kluwer Academic/Plenum Publishers; 2002. The epidemiology of HIV and AIDS; pp. 200–17. full_text.
-
- Joint United Nations program on HIV/AIDS (UNAIDS) http://www.unaids.org/en/KnowledgeCentre/HIVData/GlobalReport/2008/2008_... - PubMed
-
- Renjifo B, Essex M. In: AIDS in Africa. 2. Essex M, Mboup S, Kanki PJ, Marlink PJ, Tlou SD, editor. New York. Kluwer Academic/Plenum Publishers; 2002. HIV-1 subtypes and recombinants; pp. 138–57. full_text.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

