Effect of hyperlipidemia on the expression of circadian genes in apolipoprotein E knock-out atherosclerotic mice
- PMID: 20040117
- PMCID: PMC2807425
- DOI: 10.1186/1476-511X-8-60
Effect of hyperlipidemia on the expression of circadian genes in apolipoprotein E knock-out atherosclerotic mice
Abstract
Background: Circadian patterns of cardiovascular vulnerability were well characterized, with a peak incidence of acute myocardial infarction and stroke secondary to atherosclerosis in the morning, which showed the circadian clock may take part in the pathological process of atherosclerosis induced by hyperlipidemia. Hence, the effect of hyperlipidemia on the expression of circadian genes was investigated in atherosclerotic mouse model.
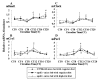
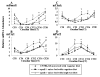
Results: In apoE-/-mice on regular chow or high-fat diet, an atherosclerotic mouse model induced by heperlipidemia, we found that the peak concentration of serum lipids was showed four or eight hours later in apoE-/- mice, compared to C57BL/6J mice. During the artificial light period, a reduce in circulating level of serum lipids corresponded with the observed increase of the expression levels of some the transcription factors involved in lipid metabolism, such as PPARalpha and RXRalpha. Meanwhile, the expression of circadian genes was changed following with amplitude reduced or the peak mRNA level delayed.
Conclusions: Our studies indicated that heperlipidemia altered both the rhythmicity and expression of circadian genes. Diet-induced circadian disruption may affect the process of atherosclerosis and some acute cardiovascular disease.
Figures
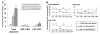




Similar articles
-
A novel BET bromodomain inhibitor, RVX-208, shows reduction of atherosclerosis in hyperlipidemic ApoE deficient mice.Atherosclerosis. 2014 Sep;236(1):91-100. doi: 10.1016/j.atherosclerosis.2014.06.008. Epub 2014 Jun 28. Atherosclerosis. 2014. PMID: 25016363
-
Urotensin II receptor knockout mice on an ApoE knockout background fed a high-fat diet exhibit an enhanced hyperlipidemic and atherosclerotic phenotype.Circ Res. 2009 Sep 25;105(7):686-95, 19 p following 695. doi: 10.1161/CIRCRESAHA.107.168799. Epub 2009 Aug 20. Circ Res. 2009. PMID: 19696412
-
Hawthorn (Crataegus pinnatifida Bunge) leave flavonoids attenuate atherosclerosis development in apoE knock-out mice.J Ethnopharmacol. 2017 Feb 23;198:479-488. doi: 10.1016/j.jep.2017.01.040. Epub 2017 Jan 21. J Ethnopharmacol. 2017. PMID: 28119096
-
Rhythm changes of clock genes, apoptosis-related genes and atherosclerosis-related genes in apolipoprotein E knockout mice.Can J Cardiol. 2009 Aug;25(8):473-9. doi: 10.1016/s0828-282x(09)70122-9. Can J Cardiol. 2009. PMID: 19668782 Free PMC article.
-
Progress and challenges in translating the biology of atherosclerosis.Nature. 2011 May 19;473(7347):317-25. doi: 10.1038/nature10146. Nature. 2011. PMID: 21593864 Review.
Cited by
-
Altered Clock and Lipid Metabolism-Related Genes in Atherosclerotic Mice Kept with Abnormal Lighting Condition.Biomed Res Int. 2016;2016:5438589. doi: 10.1155/2016/5438589. Epub 2016 Aug 18. Biomed Res Int. 2016. PMID: 27631008 Free PMC article.
-
The Effect of Diet on the Cardiac Circadian Clock in Mice: A Systematic Review.Metabolites. 2022 Dec 15;12(12):1273. doi: 10.3390/metabo12121273. Metabolites. 2022. PMID: 36557311 Free PMC article. Review.
-
Circadian Rhythm Alteration of the Core Clock Genes and the Lipid Metabolism Genes Induced by High-Fat Diet (HFD) in the Liver Tissue of the Chinese Soft-Shelled Turtle (Trionyx sinensis).Genes (Basel). 2024 Jan 25;15(2):157. doi: 10.3390/genes15020157. Genes (Basel). 2024. PMID: 38397147 Free PMC article.
-
Circadian Mechanisms in Brain Fluid Biology.Circ Res. 2024 Mar 15;134(6):711-726. doi: 10.1161/CIRCRESAHA.123.323516. Epub 2024 Mar 14. Circ Res. 2024. PMID: 38484035 Free PMC article. Review.
-
Degeneration and energy shortage in the suprachiasmatic nucleus underlies the circadian rhythm disturbance in ApoE-/- mice: implications for Alzheimer's disease.Sci Rep. 2016 Nov 8;6:36335. doi: 10.1038/srep36335. Sci Rep. 2016. PMID: 27824104 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

