The nuclear experience of CPEB: implications for RNA processing and translational control
- PMID: 20040591
- PMCID: PMC2811663
- DOI: 10.1261/rna.1779810
The nuclear experience of CPEB: implications for RNA processing and translational control
Abstract
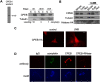
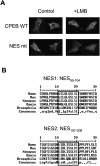
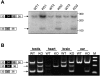
CPEB is a sequence-specific RNA binding protein that promotes polyadenylation-induced translation in early development, during cell cycle progression and cellular senescence, and following neuronal synapse stimulation. It controls polyadenylation and translation through other interacting molecules, most notably the poly(A) polymerase Gld2, the deadenylating enzyme PARN, and the eIF4E-binding protein Maskin. Here, we report that CPEB shuttles between the nucleus and cytoplasm and that its export occurs via the CRM1-dependent pathway. In the nucleus of Xenopus oocytes, CPEB associates with lampbrush chromosomes and several proteins involved in nuclear RNA processing. CPEB also interacts with Maskin in the nucleus as well as with CPE-containing mRNAs. Although the CPE does not regulate mRNA export, it influences the degree to which mRNAs are translationally repressed in the cytoplasm. Moreover, CPEB directly or indirectly mediates the alternative splicing of at least one pre-mRNA in mouse embryo fibroblasts as well as certain mouse tissues. We propose that CPEB, together with Maskin, binds mRNA in the nucleus to ensure tight translational repression upon export to the cytoplasm. In addition, we propose that nuclear CPEB regulates specific pre-mRNA alternative splicing.
Figures


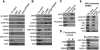



References
-
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. - PubMed
-
- Bouvet P, Wolffe AP. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell. 1994;77:931–941. - PubMed
-
- Braddock M, Thorburn AM, Chambers A, Elliott GD, Anderson GJ, Kingsman AJ, Kingsman SM. A nuclear translational block imposed by the HIV-1 U3 region is relieved by the Tat-TAR interaction. Cell. 1990;62:1123–1133. - PubMed
-
- Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16:107–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
