Discovery of novel proteasome inhibitors using a high-content cell-based screening system
- PMID: 20041034
- PMCID: PMC2797363
- DOI: 10.1371/journal.pone.0008503
Discovery of novel proteasome inhibitors using a high-content cell-based screening system
Abstract
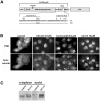
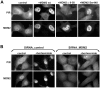
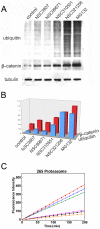
The regulated degradation of damaged or misfolded proteins, as well as down-regulation of key signaling proteins, within eukaryotic and bacterial cells is catalyzed primarily by large, ATP-dependent multimeric proteolytic complexes, termed proteasomes. Inhibition of proteasomal activity affects a wide variety of physiological and pathological processes, and was found to be particularly effective for cancer therapy. We report here on the development of a novel high throughput assay for proteasome inhibition using a unique, highly sensitive live-cell screening, based on the cytoplasm-to-nucleus translocation of a fluorescent proteasome inhibition reporter (PIR) protein, consisting of nuclear localization signal-deficient p53 derivative. We further show here that mdm2, a key negative regulator of p53 plays a key role in the accumulation of PIR in the nucleus upon proteasome inhibition. Using this assay, we have screened the NCI Diversity Set library, containing 1,992 low molecular weight synthetic compounds, and identified four proteasome inhibitors. The special features of the current screen, compared to those of other approaches are discussed.
Conflict of interest statement
Figures
Similar articles
-
A cell-based high-throughput assay for the screening of small-molecule inhibitors of p53-MDM2 interaction.J Biomol Screen. 2011 Apr;16(4):450-6. doi: 10.1177/1087057111399191. J Biomol Screen. 2011. PMID: 21471462
-
Development and validation of a high-content screening assay to identify inhibitors of cytoplasmic dynein-mediated transport of glucocorticoid receptor to the nucleus.Assay Drug Dev Technol. 2012 Oct;10(5):432-56. doi: 10.1089/adt.2012.456. Epub 2012 Jul 25. Assay Drug Dev Technol. 2012. PMID: 22830992 Free PMC article.
-
Implementation of a 220,000-compound HCS campaign to identify disruptors of the interaction between p53 and hDM2 and characterization of the confirmed hits.J Biomol Screen. 2010 Aug;15(7):766-82. doi: 10.1177/1087057110375304. Epub 2010 Jul 16. J Biomol Screen. 2010. PMID: 20639499
-
Development and application of PI3K assays for novel drug discovery.Expert Opin Drug Discov. 2015 Feb;10(2):171-86. doi: 10.1517/17460441.2015.997205. Epub 2014 Dec 30. Expert Opin Drug Discov. 2015. PMID: 25547459 Review.
-
Target Discovery for New Antitubercular Drugs Using a Large Dataset of Growth Inhibitors from PubChem.Infect Disord Drug Targets. 2020;20(3):352-366. doi: 10.2174/1871526519666181205163810. Infect Disord Drug Targets. 2020. PMID: 30520384 Review.
Cited by
-
Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases.Mol Ther. 2013 Jun;21(6):1259-69. doi: 10.1038/mt.2013.65. Epub 2013 Apr 16. Mol Ther. 2013. PMID: 23587921 Free PMC article.
-
Targeting neuroblastoma stem cells with retinoic acid and proteasome inhibitor.PLoS One. 2013 Oct 7;8(10):e76761. doi: 10.1371/journal.pone.0076761. eCollection 2013. PLoS One. 2013. PMID: 24116151 Free PMC article.
-
A High-Throughput Cell-Based Screen Identified a 2-[(E)-2-Phenylvinyl]-8-Quinolinol Core Structure That Activates p53.PLoS One. 2016 Apr 28;11(4):e0154125. doi: 10.1371/journal.pone.0154125. eCollection 2016. PLoS One. 2016. PMID: 27124407 Free PMC article.
-
Synergistic activation of bat SARS-like coronaviruses spike protein by elastase and TMPRSS2.Sci Rep. 2025 Jul 21;15(1):26469. doi: 10.1038/s41598-025-11600-y. Sci Rep. 2025. PMID: 40691229 Free PMC article.
-
Gambogic acid is cytotoxic to cancer cells through inhibition of the ubiquitin-proteasome system.Invest New Drugs. 2013 Jun;31(3):587-98. doi: 10.1007/s10637-012-9902-y. Epub 2012 Nov 20. Invest New Drugs. 2013. PMID: 23179339
References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. - PubMed
-
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. - PubMed
-
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. - PubMed
-
- Orlowski M, Cardozo C, Michaud C. Evidence for the presence of five distinct proteolytic components in the pituitary multicatalytic proteinase complex. Properties of two components cleaving bonds on the carboxyl side of branched chain and small neutral amino acids. Biochemistry. 1993;32:1563–1572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

