Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers
- PMID: 20041112
- PMCID: PMC2795174
- DOI: 10.1371/journal.pone.0008451
Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers
Abstract
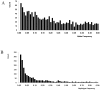
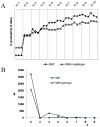
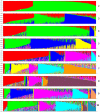
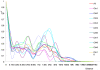
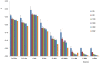
A newly developed maize Illumina GoldenGate Assay with 1536 SNPs from 582 loci was used to genotype a highly diverse global maize collection of 632 inbred lines from temperate, tropical, and subtropical public breeding programs. A total of 1229 informative SNPs and 1749 haplotypes within 327 loci was used to estimate the genetic diversity, population structure, and familial relatedness. Population structure identified tropical and temperate subgroups, and complex familial relationships were identified within the global collection. Linkage disequilibrium (LD) was measured overall and within chromosomes, allelic frequency groups, subgroups related by geographic origin, and subgroups of different sample sizes. The LD decay distance differed among chromosomes and ranged between 1 to 10 kb. The LD distance increased with the increase of minor allelic frequency (MAF), and with smaller sample sizes, encouraging caution when using too few lines in a study. The LD decay distance was much higher in temperate than in tropical and subtropical lines, because tropical and subtropical lines are more diverse and contain more rare alleles than temperate lines. A core set of inbreds was defined based on haplotypes, and 60 lines capture 90% of the haplotype diversity of the entire panel. The defined core sets and the entire collection can be used widely for different research targets.
Conflict of interest statement
Figures
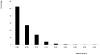
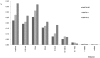
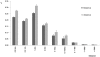
References
-
- GCDT. Global Strategy for the Ex situ Conservation and Utilization of Maize Germplasm. Global Crop Diversity Trust, Rome, Italy. 2007. http://www.croptrust.org/documents/web/Maize-Strategy-FINAL-18Sept07.pdf.
-
- Reif JC, Melchinger AE, Xia XC, Warburton ML, Hoisington DA, et al. Use of SSRs for establishing heterotic groups in subtropical maize. Theor Appl Genet. 2003;107:947–957. - PubMed
-
- Xia XC, Reif JC, Hoisington DA, Melchinger AE, Frisch M, et al. Genetic diversity among CIMMYT maize inbred lines investigated with SSR markers: I. Lowland tropical maize. Crop Sci. 2004;44:2230–2237.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

