Steered molecular dynamics simulations reveal the likelier dissociation pathway of imatinib from its targeting kinases c-Kit and Abl
- PMID: 20041122
- PMCID: PMC2795779
- DOI: 10.1371/journal.pone.0008470
Steered molecular dynamics simulations reveal the likelier dissociation pathway of imatinib from its targeting kinases c-Kit and Abl
Abstract
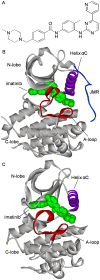
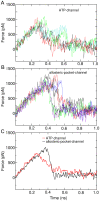
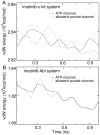
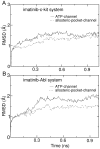
Development of small molecular kinase inhibitors has recently been the central focus in drug discovery. And type II kinase inhibitors that target inactive conformation of kinases have attracted particular attention since their potency and selectivity are thought to be easier to achieve compared with their counterpart type I inhibitors that target active conformation of kinases. Although mechanisms underlying the interactions between type II inhibitors and their targeting kinases have been widely studied, there are still some challenging problems, for example, how type II inhibitors associate with or dissociate from their targeting kinases. In this investigation, steered molecular dynamics simulations have been carried out to explore the possible dissociation pathways of typical type II inhibitor imatinib from its targeting protein kinases c-Kit and Abl. The simulation results indicate that the most favorable pathway for imatinib dissociation corresponds to the ATP-channel rather than the relatively wider allosteric-pocket-channel, which is mainly due to the different van der Waals interaction that the ligand suffers during dissociation. Nevertheless, the direct reason comes from the fact that the residues composing the ATP-channel are more flexible than that forming the allosteric-pocket-channel. The present investigation suggests that a bulky hydrophobic head is unfavorable, but a large polar tail is allowed for a potent type II inhibitor. The information obtained here can be used to direct the discovery of type II kinase inhibitors.
Conflict of interest statement
Figures




Similar articles
-
Computational analysis of the binding specificity of Gleevec to Abl, c-Kit, Lck, and c-Src tyrosine kinases.J Am Chem Soc. 2013 Oct 2;135(39):14741-53. doi: 10.1021/ja405939x. Epub 2013 Sep 20. J Am Chem Soc. 2013. PMID: 24001034 Free PMC article.
-
Conformation-selective inhibitors reveal differences in the activation and phosphate-binding loops of the tyrosine kinases Abl and Src.ACS Chem Biol. 2013 Dec 20;8(12):2734-43. doi: 10.1021/cb400663k. Epub 2013 Oct 29. ACS Chem Biol. 2013. PMID: 24106839 Free PMC article.
-
Explaining why Gleevec is a specific and potent inhibitor of Abl kinase.Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1664-9. doi: 10.1073/pnas.1214330110. Epub 2013 Jan 14. Proc Natl Acad Sci U S A. 2013. PMID: 23319661 Free PMC article.
-
Overcoming kinase resistance in chronic myeloid leukemia.Int J Biochem Cell Biol. 2008;40(3):334-43. doi: 10.1016/j.biocel.2007.10.001. Int J Biochem Cell Biol. 2008. PMID: 18401881 Review.
-
Protein kinase inhibitors.Folia Biol (Praha). 2006;52(4):137-48. Folia Biol (Praha). 2006. PMID: 17116285 Review.
Cited by
-
A Guide to In Silico Drug Design.Pharmaceutics. 2022 Dec 23;15(1):49. doi: 10.3390/pharmaceutics15010049. Pharmaceutics. 2022. PMID: 36678678 Free PMC article. Review.
-
Beyond standard molecular dynamics: investigating the molecular mechanisms of G protein-coupled receptors with enhanced molecular dynamics methods.Adv Exp Med Biol. 2014;796:95-125. doi: 10.1007/978-94-007-7423-0_6. Adv Exp Med Biol. 2014. PMID: 24158803 Free PMC article. Review.
-
Insight into resistance mechanisms of AZD4547 and E3810 to FGFR1 gatekeeper mutation via theoretical study.Drug Des Devel Ther. 2017 Feb 17;11:451-461. doi: 10.2147/DDDT.S129991. eCollection 2017. Drug Des Devel Ther. 2017. PMID: 28255231 Free PMC article.
-
In silico study of Aquaporin V: Effects and affinity of the central pore-occluding lipid.Biophys Chem. 2013 Jan;171:24-30. doi: 10.1016/j.bpc.2012.09.004. Epub 2012 Oct 2. Biophys Chem. 2013. PMID: 23176748 Free PMC article.
-
Diversity of Long-Lived Intermediates along the Binding Pathway of Imatinib to Abl Kinase Revealed by MD Simulations.J Chem Theory Comput. 2020 Dec 8;16(12):7852-7865. doi: 10.1021/acs.jctc.0c00739. Epub 2020 Nov 4. J Chem Theory Comput. 2020. PMID: 33147951 Free PMC article.
References
-
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. - PubMed
-
- Cohen P. Protein kinases - the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. - PubMed
-
- Dancey J, Sausville EA. Issues and progress with protein kinase inhibitors for cancer treatment. Nat Rev Drug Discov. 2003;2:296–313. - PubMed
-
- Noble MEM, Endicott JA, Johnson LN. Protein kinase inhibitors: Insights into drug design from structure. Science. 2004;303:1800–1805. - PubMed
-
- Thaimattam R, Banerjee R, Miglani R, Iqbal J. Protein kinase inhibitors: Structural insights into selectivity. Curr Pharm Des. 2007;13:2751–2765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

