Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
- PMID: 20041127
- PMCID: PMC2796168
- DOI: 10.1371/journal.pone.0008514
Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
Abstract
Background: Chemokines are a subset of cytokines responsible for controlling the cellular migration of inflammatory cells through interaction with seven transmembrane G protein-coupled receptors. The blocking of a chemokine-receptor interaction results in a reduced inflammatory response, and represents a possible anti-inflammatory strategy, a strategy that is already employed by some virus and parasites. Anti-chemokine activity has been described in the extracts of tick salivary glands, and we have recently described the cloning and characterization of such chemokine binding proteins from the salivary glands, which we have named Evasins.
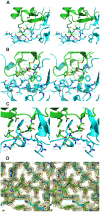
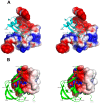
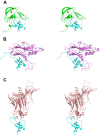
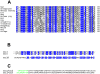
Methodology/principal findings: We have solved the structure of Evasin-1, a very small and highly selective chemokine-binding protein, by x-ray crystallography and report that the structure is novel, with no obvious similarity to the previously described structures of viral chemokine binding proteins. Moreover it does not possess a known fold. We have also solved the structure of the complex of Evasin-1 and its high affinity ligand, CCL3. The complex is a 1:1 heterodimer in which the N-terminal region of CCL3 forms numerous contacts with Evasin-1, including prominent pi-pi interactions between residues Trp89 and Phe14 of the binding protein and Phe29 and Phe13 of the chemokine.
Conclusions/significance: However, these interactions do not appear to be crucial for the selectivity of the binding protein, since these residues are found in CCL5, which is not a ligand for Evasin-1. The selectivity of the interaction would appear to lie in the N-terminal residues of the chemokine, which form the "address" whereas the hydrophobic interactions in the rest of the complex would serve primarily to stabilize the complex. A thorough understanding of the binding mode of this small protein, and its other family members, could be very informative in the design of potent neutralizing molecules of pro-inflammatory mediators of the immune system, such as chemokines.
Conflict of interest statement
Figures
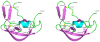
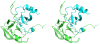
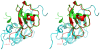
Similar articles
-
The N-terminal domain of a tick evasin is critical for chemokine binding and neutralization and confers specific binding activity to other evasins.J Biol Chem. 2018 Apr 20;293(16):6134-6146. doi: 10.1074/jbc.RA117.000487. Epub 2018 Feb 27. J Biol Chem. 2018. PMID: 29487134 Free PMC article.
-
Identification of the pharmacophore of the CC chemokine-binding proteins Evasin-1 and -4 using phage display.J Biol Chem. 2014 Nov 14;289(46):31846-31855. doi: 10.1074/jbc.M114.599233. Epub 2014 Sep 29. J Biol Chem. 2014. PMID: 25266725 Free PMC article.
-
Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick Rhipicephalus sanguineus.J Biol Chem. 2007 Sep 14;282(37):27250-27258. doi: 10.1074/jbc.M704706200. Epub 2007 Jul 19. J Biol Chem. 2007. PMID: 17640866
-
Using evasins to target the chemokine network in inflammation.Adv Protein Chem Struct Biol. 2020;119:1-38. doi: 10.1016/bs.apcsb.2019.09.003. Epub 2019 Nov 26. Adv Protein Chem Struct Biol. 2020. PMID: 31997766 Review.
-
Evasins: Tick Salivary Proteins that Inhibit Mammalian Chemokines.Trends Biochem Sci. 2020 Feb;45(2):108-122. doi: 10.1016/j.tibs.2019.10.003. Epub 2019 Nov 1. Trends Biochem Sci. 2020. PMID: 31679840 Free PMC article. Review.
Cited by
-
The binding and specificity of chemokine binding proteins, through the lens of experiment and computation.Biomed J. 2022 Jun;45(3):439-453. doi: 10.1016/j.bj.2021.07.004. Epub 2021 Jul 24. Biomed J. 2022. PMID: 34311129 Free PMC article. Review.
-
Ticks from diverse genera encode chemokine-inhibitory evasin proteins.J Biol Chem. 2017 Sep 22;292(38):15670-15680. doi: 10.1074/jbc.M117.807255. Epub 2017 Aug 4. J Biol Chem. 2017. PMID: 28778927 Free PMC article.
-
The N-terminal domain of a tick evasin is critical for chemokine binding and neutralization and confers specific binding activity to other evasins.J Biol Chem. 2018 Apr 20;293(16):6134-6146. doi: 10.1074/jbc.RA117.000487. Epub 2018 Feb 27. J Biol Chem. 2018. PMID: 29487134 Free PMC article.
-
Chemokine Heteromers and Their Impact on Cellular Function-A Conceptual Framework.Int J Mol Sci. 2023 Jun 30;24(13):10925. doi: 10.3390/ijms241310925. Int J Mol Sci. 2023. PMID: 37446102 Free PMC article. Review.
-
International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors.Pharmacol Rev. 2013 Nov 11;66(1):1-79. doi: 10.1124/pr.113.007724. Print 2014. Pharmacol Rev. 2013. PMID: 24218476 Free PMC article. Review.
References
-
- Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–115. - PubMed
-
- Johnson Z, Borlat R, Wells TNC, Kosco-Vilbois MH, Proudfoot AEI. A novel strategy for blocking chemokine mediated inflammation. Inflammation Research. 2003;52:S187–S189.
-
- Schoeler GB, Wikel SK. Modulation of host immunity by haematophagous arthropods. Ann Trop Med Parasitol. 2001;95:755–771. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

