A gene expression signature of invasive potential in metastatic melanoma cells
- PMID: 20041153
- PMCID: PMC2794539
- DOI: 10.1371/journal.pone.0008461
A gene expression signature of invasive potential in metastatic melanoma cells
Abstract
Background: We are investigating the molecular basis of melanoma by defining genomic characteristics that correlate with tumour phenotype in a novel panel of metastatic melanoma cell lines. The aim of this study is to identify new prognostic markers and therapeutic targets that might aid clinical cancer diagnosis and management.
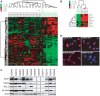
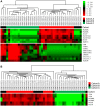
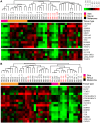
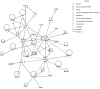
Principal findings: Global transcript profiling identified a signature featuring decreased expression of developmental and lineage specification genes including MITF, EDNRB, DCT, and TYR, and increased expression of genes involved in interaction with the extracellular environment, such as PLAUR, VCAN, and HIF1a. Migration assays showed that the gene signature correlated with the invasive potential of the cell lines, and external validation by using publicly available data indicated that tumours with the invasive gene signature were less melanocytic and may be more aggressive. The invasion signature could be detected in both primary and metastatic tumours suggesting that gene expression conferring increased invasive potential in melanoma may occur independently of tumour stage.
Conclusions: Our data supports the hypothesis that differential developmental gene expression may drive invasive potential in metastatic melanoma, and that melanoma heterogeneity may be explained by the differing capacity of melanoma cells to both withstand decreased expression of lineage specification genes and to respond to the tumour microenvironment. The invasion signature may provide new possibilities for predicting which primary tumours are more likely to metastasize, and which metastatic tumours might show a more aggressive clinical course.
Conflict of interest statement
Figures

References
-
- Hocker TL, Singh MK, Tsao H. Melanoma genetics and therapeutic approaches in the 21st century: moving from the benchside to the bedside. J Invest Dermatol. 2008;128:2575–2595. - PubMed
-
- Francken AB, Accortt NA, Shaw HM, Wiener M, Soong SJ, et al. Prognosis and determinants of outcome following locoregional or distant recurrence in patients with cutaneous melanoma. Ann Surg Oncol. 2008;15:1476–1484. - PubMed
-
- Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19:3622–3634. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

