Identification and developmental expression of Xenopus laevis SUMO proteases
- PMID: 20041154
- PMCID: PMC2794540
- DOI: 10.1371/journal.pone.0008462
Identification and developmental expression of Xenopus laevis SUMO proteases
Abstract
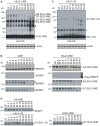
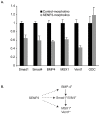
SUMO proteins are small ubiquitin-related modifiers. All SUMOs are synthesized as propeptides that are post-translationally cleaved prior to conjugation. After processing, SUMOs become covalently conjugated to cellular targets through a pathway that is similar to ubiquitination. Ubiquitin like protein proteases/Sentrin specific proteases (Ulp/SENPs) mediate both processing and deconjugation of SUMOs. The action of Ulp/SENPs makes SUMOylation a highly dynamic post-translational modification. To investigate how Ulp/SENPs are regulated in a developmental context, we isolated and characterized all Ulp/SENPs in Xenopus laevis. Xenopus possess homologues of mammalian SENP3, 5, 6 and 7. All of these enzymes reacted with HA-tagged vinyl sulfone derivatives of SUMO-2 (HA-SU2-VS) but not SUMO-1 (HA-SU1-VS), suggesting that they act primarily on SUMO-2 and -3. In contrast, Xenopus possess a single member of the SENP1/SENP2 subfamily of Ulp/SENPs, most closely related to mammalian SENP1. Xenopus SENP1 reacted with HA-SU1-VS and HA-SU2-VS, suggesting that it acts on all SUMO paralogues. We analyzed the mRNA and protein levels for each of the Ulp/SENPs through development; we found that they show distinct patterns of expression that may involve both transcriptional and post-transcriptional regulation. Finally, we have characterized the developmental function of the most abundant Ulp/SENP found within Xenopus eggs, SENP3. Depletion of SENP3 using morpholino antisense oligonucleotides (morpholinos) caused accumulation of high molecular weight SUMO-2/3 conjugated species, defects in developing embryos and changes in the expression of some genes regulated by the transforming growth factor beta (TGF-beta) pathway. These findings collectively indicate that SUMO proteases are both highly regulated and essential for normal development.
Conflict of interest statement
Figures



Similar articles
-
Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases.J Cell Biol. 2008 Nov 17;183(4):589-95. doi: 10.1083/jcb.200807185. J Cell Biol. 2008. PMID: 19015314 Free PMC article.
-
SUMO proteases as potential targets for cancer therapy.Postepy Hig Med Dosw (Online). 2017 Dec 8;71(0):997-1004. doi: 10.5604/01.3001.0010.6667. Postepy Hig Med Dosw (Online). 2017. PMID: 29225200 Review.
-
Modification in reverse: the SUMO proteases.Trends Biochem Sci. 2007 Jun;32(6):286-95. doi: 10.1016/j.tibs.2007.05.002. Epub 2007 May 17. Trends Biochem Sci. 2007. PMID: 17499995 Review.
-
Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs.J Biol Chem. 2007 Sep 7;282(36):26217-24. doi: 10.1074/jbc.M702444200. Epub 2007 Jun 25. J Biol Chem. 2007. PMID: 17591783
-
XSENP1, a novel sumo-specific protease in Xenopus, inhibits normal head formation by down-regulation of Wnt/beta-catenin signalling.Genes Cells. 2004 Aug;9(8):723-36. doi: 10.1111/j.1356-9597.2004.00757.x. Genes Cells. 2004. PMID: 15298680
Cited by
-
Starting and stopping SUMOylation. What regulates the regulator?Chromosoma. 2013 Dec;122(6):451-63. doi: 10.1007/s00412-013-0422-0. Epub 2013 Jun 28. Chromosoma. 2013. PMID: 23812602 Review.
-
The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase.Cell Cycle. 2010 Aug 15;9(16):3194-201. doi: 10.4161/cc.9.16.12619. Epub 2010 Aug 9. Cell Cycle. 2010. PMID: 20724819 Free PMC article. Review.
-
The SUMO protease SENP6 is essential for inner kinetochore assembly.J Cell Biol. 2010 Mar 8;188(5):681-92. doi: 10.1083/jcb.200909008. J Cell Biol. 2010. PMID: 20212317 Free PMC article.
-
Distribution and paralogue specificity of mammalian deSUMOylating enzymes.Biochem J. 2010 Sep 1;430(2):335-44. doi: 10.1042/BJ20100504. Biochem J. 2010. PMID: 20590526 Free PMC article.
-
SUMO2/3 modification of cyclin E contributes to the control of replication origin firing.Nat Commun. 2013;4:1850. doi: 10.1038/ncomms2875. Nat Commun. 2013. PMID: 23673635 Free PMC article.
References
-
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. - PubMed
-
- Hay RT. Role of ubiquitin-like proteins in transcriptional regulation. Ernst Schering Res Found Workshop. 2006:173–192. - PubMed
-
- Seeler JS, Bischof O, Nacerddine K, Dejean A. SUMO, the three Rs and cancer. Curr Top Microbiol Immunol. 2007;313:49–71. - PubMed
-
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. - PubMed
-
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

