A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus
- PMID: 20041173
- PMCID: PMC2791863
- DOI: 10.1371/journal.ppat.1000705
A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus
Abstract
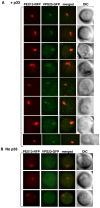
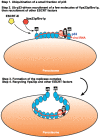
Plus-stranded RNA viruses replicate in infected cells by assembling viral replicase complexes consisting of viral- and host-coded proteins. Previous genome-wide screens with Tomato bushy stunt tombusvirus (TBSV) in a yeast model host revealed the involvement of seven ESCRT (endosomal sorting complexes required for transport) proteins in viral replication. In this paper, we show that the expression of dominant negative Vps23p, Vps24p, Snf7p, and Vps4p ESCRT factors inhibited virus replication in the plant host, suggesting that tombusviruses co-opt selected ESCRT proteins for the assembly of the viral replicase complex. We also show that TBSV p33 replication protein interacts with Vps23p ESCRT-I and Bro1p accessory ESCRT factors. The interaction with p33 leads to the recruitment of Vps23p to the peroxisomes, the sites of TBSV replication. The viral replicase showed reduced activity and the minus-stranded viral RNA in the replicase became more accessible to ribonuclease when derived from vps23Delta or vps24Delta yeast, suggesting that the protection of the viral RNA is compromised within the replicase complex assembled in the absence of ESCRT proteins. The recruitment of ESCRT proteins is needed for the precise assembly of the replicase complex, which might help the virus evade recognition by the host defense surveillance system and/or prevent viral RNA destruction by the gene silencing machinery.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





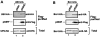
References
-
- Nagy PD. Yeast as a model host to explore plant virus-host interactions. Annu Rev Phytopathol. 2008;46:217–242. - PubMed
-
- Noueiry AO, Ahlquist P. Brome Mosaic Virus RNA Replication: Revealing the Role of the Host in RNA Virus Replication. Annu Rev Phytopathol 2003 - PubMed
-
- Nagy PD, Pogany J. Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol Biol. 2008;451:55–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

