Autoimmune disease classification by inverse association with SNP alleles
- PMID: 20041220
- PMCID: PMC2791168
- DOI: 10.1371/journal.pgen.1000792
Autoimmune disease classification by inverse association with SNP alleles
Abstract
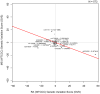
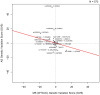
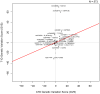
With multiple genome-wide association studies (GWAS) performed across autoimmune diseases, there is a great opportunity to study the homogeneity of genetic architectures across autoimmune disease. Previous approaches have been limited in the scope of their analysis and have failed to properly incorporate the direction of allele-specific disease associations for SNPs. In this work, we refine the notion of a genetic variation profile for a given disease to capture strength of association with multiple SNPs in an allele-specific fashion. We apply this method to compare genetic variation profiles of six autoimmune diseases: multiple sclerosis (MS), ankylosing spondylitis (AS), autoimmune thyroid disease (ATD), rheumatoid arthritis (RA), Crohn's disease (CD), and type 1 diabetes (T1D), as well as five non-autoimmune diseases. We quantify pair-wise relationships between these diseases and find two broad clusters of autoimmune disease where SNPs that make an individual susceptible to one class of autoimmune disease also protect from diseases in the other autoimmune class. We find that RA and AS form one such class, and MS and ATD another. We identify specific SNPs and genes with opposite risk profiles for these two classes. We furthermore explore individual SNPs that play an important role in defining similarities and differences between disease pairs. We present a novel, systematic, cross-platform approach to identify allele-specific relationships between disease pairs based on genetic variation as well as the individual SNPs which drive the relationships. While recognizing similarities between diseases might lead to identifying novel treatment options, detecting differences between diseases previously thought to be similar may point to key novel disease-specific genes and pathways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Thorsby E, Lie BA. HLA associated genetic predisposition to autoimmune diseases: Genes involved and possible mechanisms. Transpl Immunol. 2005;14:175–182. - PubMed
-
- Zanelli E, Breedveld FC, de Vries RR. HLA association with autoimmune disease: a failure to protect? Rheumatology (Oxford) 2000;39:1060–1066. - PubMed
-
- Robinson D, Jr, Hackett M, Wong J, Kimball AB, Cohen R, et al. Co-occurrence and comorbidities in patients with immune-mediated inflammatory disorders: an exploration using US healthcare claims data, 2001–2002. Curr Med Res Opin. 2006;22:989–1000. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

