Quantitative analysis of replication-related mutation and selection pressures in bacterial chromosomes and plasmids using generalised GC skew index
- PMID: 20042086
- PMCID: PMC2804667
- DOI: 10.1186/1471-2164-10-640
Quantitative analysis of replication-related mutation and selection pressures in bacterial chromosomes and plasmids using generalised GC skew index
Abstract
Background: Due to their bi-directional replication machinery starting from a single finite origin, bacterial genomes show characteristic nucleotide compositional bias between the two replichores, which can be visualised through GC skew or (C-G)/(C+G). Although this polarisation is used for computational prediction of replication origins in many bacterial genomes, the degree of GC skew visibility varies widely among different species, necessitating a quantitative measurement of GC skew strength in order to provide confidence measures for GC skew-based predictions of replication origins.
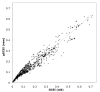
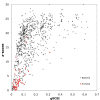
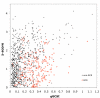
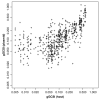
Results: Here we discuss a quantitative index for the measurement of GC skew strength, named the generalised GC skew index (gGCSI), which is applicable to genomes of any length, including bacterial chromosomes and plasmids. We demonstrate that gGCSI is independent of the window size and can thus be used to compare genomes with different sizes, such as bacterial chromosomes and plasmids. It can suggest the existence of different replication mechanisms in archaea and of rolling-circle replication in plasmids. Correlation of gGCSI values between plasmids and their corresponding host chromosomes suggests that within the same strain, these replicons have reproduced using the same replication machinery and thus exhibit similar strengths of replication strand skew.
Conclusions: gGCSI can be applied to genomes of any length and thus allows comparative study of replication-related mutation and selection pressures in genomes of different lengths such as bacterial chromosomes and plasmids. Using gGCSI, we showed that replication-related mutation or selection pressure is similar for replicons with similar machinery.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

