Virus infection elevates transcriptional activity of miR164a promoter in plants
- PMID: 20042107
- PMCID: PMC2809068
- DOI: 10.1186/1471-2229-9-152
Virus infection elevates transcriptional activity of miR164a promoter in plants
Abstract
Background: Micro RNAs (miRs) constitute a large group of endogenous small RNAs that have crucial roles in many important plant functions. Virus infection and transgenic expression of viral proteins alter accumulation and activity of miRs and so far, most of the published evidence involves post-transcriptional regulations.
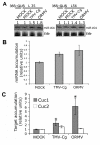
Results: Using transgenic plants expressing a reporter gene under the promoter region of a characterized miR (P-miR164a), we monitored the reporter gene expression in different tissues and during Arabidopsis development. Strong expression was detected in both vascular tissues and hydathodes. P-miR164a activity was developmentally regulated in plants with a maximum expression at stages 1.12 to 5.1 (according to Boyes, 2001) along the transition from vegetative to reproductive growth. Upon quantification of P-miR164a-derived GUS activity after Tobacco mosaic virus Cg or Oilseed rape mosaic virus (ORMV) infection and after hormone treatments, we demonstrated that ORMV and gibberellic acid elevated P-miR164a activity. Accordingly, total mature miR164, precursor of miR164a and CUC1 mRNA (a miR164 target) levels increased after virus infection and interestingly the most severe virus (ORMV) produced the strongest promoter induction.
Conclusion: This work shows for the first time that the alteration of miR pathways produced by viral infections possesses a transcriptional component. In addition, the degree of miR alteration correlates with virus severity since a more severe virus produces a stronger P-miR164a induction.
Figures



References
-
- Xie Z, Qi X. Diverse small RNA-directed silencing pathways in plants. Biochimica et biophysica acta. 2008;1779:720–724. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

