Kinetics and intracellular location of intramolecular disulfide bond formation mediated by the cytoplasmic redox system encoded by vaccinia virus
- PMID: 20042211
- PMCID: PMC2823943
- DOI: 10.1016/j.virol.2009.11.026
Kinetics and intracellular location of intramolecular disulfide bond formation mediated by the cytoplasmic redox system encoded by vaccinia virus
Abstract
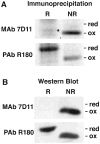
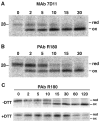
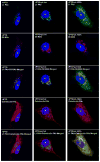
Poxviruses encode a redox system for intramolecular disulfide bond formation in cytoplasmic domains of viral proteins. Our objectives were to determine the kinetics and intracellular location of disulfide bond formation. The vaccinia virus L1 myristoylated membrane protein, used as an example, has three intramolecular disulfide bonds. Reduced and disulfide-bonded forms of L1 were distinguished by electrophoretic mobility and reactivity with monoclonal and polyclonal antibodies. Because disulfide bonds formed during 5 min pulse labeling with radioactive amino acids, a protocol was devised in which dithiothreitol was present at this step. Disulfide bond formation was detected by 2 min after removal of reducing agent and was nearly complete in 10 min. When the penultimate glycine residue was mutated to prevent myristoylation, L1 was mistargeted to the endoplasmic reticulum and disulfide bond formation failed to occur. These data suggested that viral membrane association was required for oxidation of L1, providing specificity for the process.
Published by Elsevier Inc.
Figures



Similar articles
-
Vaccinia virus G4L glutaredoxin is an essential intermediate of a cytoplasmic disulfide bond pathway required for virion assembly.J Virol. 2002 Jan;76(2):467-72. doi: 10.1128/jvi.76.2.467-472.2002. J Virol. 2002. PMID: 11752136 Free PMC article.
-
Vaccinia virus A28L gene encodes an essential protein component of the virion membrane with intramolecular disulfide bonds formed by the viral cytoplasmic redox pathway.J Virol. 2004 Mar;78(5):2348-56. doi: 10.1128/jvi.78.5.2348-2356.2004. J Virol. 2004. PMID: 14963131 Free PMC article.
-
An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins.J Cell Biol. 1999 Jan 25;144(2):267-79. doi: 10.1083/jcb.144.2.267. J Cell Biol. 1999. PMID: 9922453 Free PMC article.
-
From structure to redox: The diverse functional roles of disulfides and implications in disease.Proteomics. 2017 Mar;17(6):10.1002/pmic.201600391. doi: 10.1002/pmic.201600391. Proteomics. 2017. PMID: 28044432 Free PMC article. Review.
-
Genomics perspective on disulfide bond formation.Antioxid Redox Signal. 2003 Aug;5(4):397-402. doi: 10.1089/152308603768295131. Antioxid Redox Signal. 2003. PMID: 13678527 Review.
Cited by
-
Vaccinia virus L2 protein associates with the endoplasmic reticulum near the growing edge of crescent precursors of immature virions and stabilizes a subset of viral membrane proteins.J Virol. 2011 Dec;85(23):12431-41. doi: 10.1128/JVI.05573-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917978 Free PMC article.
-
Poxvirus cell entry: how many proteins does it take?Viruses. 2012 May;4(5):688-707. doi: 10.3390/v4050688. Epub 2012 Apr 27. Viruses. 2012. PMID: 22754644 Free PMC article. Review.
-
The myristate moiety and amino terminus of vaccinia virus l1 constitute a bipartite functional region needed for entry.J Virol. 2012 May;86(10):5437-51. doi: 10.1128/JVI.06703-11. Epub 2012 Mar 7. J Virol. 2012. PMID: 22398293 Free PMC article.
-
Chemistry and Enzymology of Disulfide Cross-Linking in Proteins.Chem Rev. 2018 Feb 14;118(3):1169-1198. doi: 10.1021/acs.chemrev.7b00123. Epub 2017 Jul 12. Chem Rev. 2018. PMID: 28699750 Free PMC article. Review.
-
Characterization and structure of the vaccinia virus NF-κB antagonist A46.J Biol Chem. 2014 Feb 7;289(6):3749-62. doi: 10.1074/jbc.M113.512756. Epub 2013 Dec 19. J Biol Chem. 2014. PMID: 24356965 Free PMC article.
References
-
- Aldaz-Carroll L, Whitbeck JC, Ponce de Leon M, Lou H, Pannell LK, Lebowitz J, Fogg C, White C, Moss B, Cohen GH, Eisenberg RJ. Physical and immunological characterization of a recombinant secreted form of the membrane protein encoded by the vaccinia virus L1R gene. Virology. 2005;341:59–71. - PubMed
-
- Condit RC, Moussatche N, Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv Virus Res. 2006;66:31–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

