Evidence for multiple recent host species shifts among the Ranaviruses (family Iridoviridae)
- PMID: 20042506
- PMCID: PMC2826071
- DOI: 10.1128/JVI.01991-09
Evidence for multiple recent host species shifts among the Ranaviruses (family Iridoviridae)
Abstract
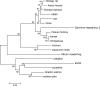
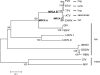
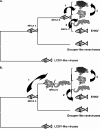
Members of the genus Ranavirus (family Iridoviridae) have been recognized as major viral pathogens of cold-blooded vertebrates. Ranaviruses have been associated with amphibians, fish, and reptiles. At this time, the relationships between ranavirus species are still unclear. Previous studies suggested that ranaviruses from salamanders are more closely related to ranaviruses from fish than they are to ranaviruses from other amphibians, such as frogs. Therefore, to gain a better understanding of the relationships among ranavirus isolates, the genome of epizootic hematopoietic necrosis virus (EHNV), an Australian fish pathogen, was sequenced. Our findings suggest that the ancestral ranavirus was a fish virus and that several recent host shifts have taken place, with subsequent speciation of viruses in their new hosts. The data suggesting several recent host shifts among ranavirus species increase concern that these pathogens of cold-blooded vertebrates may have the capacity to cross numerous poikilothermic species barriers and the potential to cause devastating disease in their new hosts.
Figures




Similar articles
-
Iridovirus infections in finfish - critical review with emphasis on ranaviruses.J Fish Dis. 2010 Feb;33(2):95-122. doi: 10.1111/j.1365-2761.2009.01110.x. Epub 2009 Dec 28. J Fish Dis. 2010. PMID: 20050967 Review.
-
Rapid differentiation of Australian, European and American ranaviruses based on variation in major capsid protein gene sequence.Mol Cell Probes. 2002 Apr;16(2):137-51. doi: 10.1006/mcpr.2001.0400. Mol Cell Probes. 2002. PMID: 12030764
-
Genomic sequence of a ranavirus (family Iridoviridae) associated with salamander mortalities in North America.Virology. 2003 Nov 10;316(1):90-103. doi: 10.1016/j.virol.2003.08.001. Virology. 2003. PMID: 14599794
-
Ranaviruses (family Iridoviridae): emerging cold-blooded killers.Arch Virol. 2002 Mar;147(3):447-70. doi: 10.1007/s007050200000. Arch Virol. 2002. PMID: 11958449 Review.
-
Ranaviruses and other members of the family Iridoviridae: Their place in the virosphere.Virology. 2017 Nov;511:259-271. doi: 10.1016/j.virol.2017.06.007. Epub 2017 Jun 23. Virology. 2017. PMID: 28648249 Review.
Cited by
-
Rana grylio virus 43R encodes an envelope protein involved in virus entry.Virus Genes. 2018 Dec;54(6):779-791. doi: 10.1007/s11262-018-1606-8. Epub 2018 Nov 8. Virus Genes. 2018. PMID: 30411182
-
Divergent transcriptomic responses underlying the ranaviruses-amphibian interaction processes on interspecies infection of Chinese giant salamander.BMC Genomics. 2018 Mar 20;19(1):211. doi: 10.1186/s12864-018-4596-y. BMC Genomics. 2018. PMID: 29558886 Free PMC article.
-
Phylogeny and differentiation of reptilian and amphibian ranaviruses detected in Europe.PLoS One. 2015 Feb 23;10(2):e0118633. doi: 10.1371/journal.pone.0118633. eCollection 2015. PLoS One. 2015. PMID: 25706285 Free PMC article.
-
Characterization of a virulent ranavirus isolated from marine ornamental fish in India.Virusdisease. 2017 Dec;28(4):373-382. doi: 10.1007/s13337-017-0408-2. Epub 2017 Nov 14. Virusdisease. 2017. PMID: 29291228 Free PMC article.
-
The Insights of Genomic Synteny and Codon Usage Preference on Genera Demarcation of Iridoviridae Family.Front Microbiol. 2021 Mar 31;12:657887. doi: 10.3389/fmicb.2021.657887. eCollection 2021. Front Microbiol. 2021. PMID: 33868215 Free PMC article.
References
-
- Ahne, W., M. Bearzotti, M. Bremont, and S. Essbauer. 1989. Comparison of European systemic piscine and amphibian iridoviruses with epizootic haematopoietic necrosis virus and frog virus 3. Zentralbl. Veterinarmed. B 45:373-383. - PubMed
-
- Ahne, W., M. Bremont, R. P. Hedrick, A. D. Hyatt, and R. J. Whittington. 1997. Iridoviruses associated with epizootic haematopoietic necrosis (EHN) in aquaculture. World J. Microbiol. Biotechnol. 13:367-373.
-
- Allender, M. C., M. M. Fry, A. R. Irizarry, L. Craig, A. J. Johnson, and M. Jones. 2006. Intracytoplasmic inclusions in circulating leukocytes from an eastern box turtle (Terrapene carolina carolina) with iridoviral infection. J. Wildl. Dis. 42:677-684. - PubMed
-
- Binder, S., A. M. Levitt, J. J. Sacks, and J. M. Hughes. 1999. Emerging infectious diseases: public health issues for the 21st century. Science 284:1311-1313. - PubMed
-
- Bloch, B., and J. L. Larsen. 1993. An iridovirus-like agent associated with systemic infection in cultured turbot Scophthalmus-maximus fry in Denmark. Dis. Aquat. Organ. 15:235-240.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

