Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection
- PMID: 20042588
- PMCID: PMC4321756
- DOI: 10.4049/jimmunol.0903058
Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection
Abstract
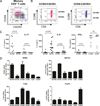
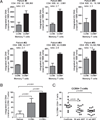
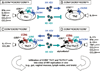
There is limited knowledge on the identity of primary CD4(+) T cell subsets selectively targeted by HIV-1 in vivo. In this study, we established a link between HIV permissiveness, phenotype/homing potential, and lineage commitment in primary CD4(+) T cells. CCR4(+)CCR6(+), CCR4(+)CCR6(-), CXCR3(+)CCR6(+), and CXCR3(+)CCR6(-) T cells expressed cytokines and transcription factors specific for Th17, Th2, Th1Th17, and Th1 lineages, respectively. CCR4(+)CCR6(+) and CXCR3(+)CCR6(+) T cells expressed the HIV coreceptors CCR5 and CXCR4 and were permissive to R5 and X4 HIV replication. CCR4(+)CCR6(-) T cells expressed CXCR4 but not CCR5 and were permissive to X4 HIV only. CXCR3(+)CCR6(-) T cells expressed CCR5 and CXCR4 but were relatively resistant to R5 and X4 HIV in vitro. Total CCR6(+) T cells compared with CCR6(-) T cells harbored higher levels of integrated HIV DNA in treatment-naive HIV-infected subjects. The frequency of total CCR6(+) T cells and those of CCR4(+)CCR6(+) and CXCR3(+)CCR6(+) T cells were diminished in chronically infected HIV-positive subjects, despite viral-suppressive therapy. A high-throughput analysis of cytokine profiles identified CXCR3(+)CCR6(+) T cells as a major source of TNF-alpha and CCL20 and demonstrated a decreased TNF-alpha/IL-10 ratio in CXCR3(+)CCR6(-) T cells. Finally, CCR4(+)CCR6(+) and CXCR3(+)CCR6(+) T cells exhibited gut- and lymph node-homing potential. Thus, we identified CCR4(+)CCR6(+) and CXCR3(+)CCR6(+) T cells as highly permissive to HIV replication, with potential to infiltrate and recruit more CCR6(+) T cells into anatomic sites of viral replication. It is necessary that new therapeutic strategies against HIV interfere with viral replication/persistence in discrete CCR6(+) T cell subsets.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures




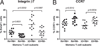
Similar articles
-
New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy.Retrovirology. 2016 Aug 24;13(1):59. doi: 10.1186/s12977-016-0293-6. Retrovirology. 2016. PMID: 27553844 Free PMC article.
-
Identification of novel HIV-1 dependency factors in primary CCR4(+)CCR6(+)Th17 cells via a genome-wide transcriptional approach.Retrovirology. 2015 Dec 10;12:102. doi: 10.1186/s12977-015-0226-9. Retrovirology. 2015. PMID: 26654242 Free PMC article.
-
Persistence of integrated HIV DNA in CXCR3 + CCR6 + memory CD4+ T cells in HIV-infected individuals on antiretroviral therapy.AIDS. 2016 Jun 19;30(10):1511-20. doi: 10.1097/QAD.0000000000001029. AIDS. 2016. PMID: 26807971 Free PMC article.
-
Migration and function of Th17 cells.Inflamm Allergy Drug Targets. 2009 Jul;8(3):221-8. doi: 10.2174/187152809788681001. Inflamm Allergy Drug Targets. 2009. PMID: 19601882 Review.
-
The Role of Immunomodulatory Receptors in the Pathogenesis of HIV Infection: A Therapeutic Opportunity for HIV Cure?Front Immunol. 2020 Jul 2;11:1223. doi: 10.3389/fimmu.2020.01223. eCollection 2020. Front Immunol. 2020. PMID: 32714317 Free PMC article. Review.
Cited by
-
Human Th17 Cells Lack HIV-Inhibitory RNases and Are Highly Permissive to Productive HIV Infection.J Virol. 2016 Aug 12;90(17):7833-47. doi: 10.1128/JVI.02869-15. Print 2016 Sep 1. J Virol. 2016. PMID: 27334595 Free PMC article.
-
Identification of CD98 as a Novel Biomarker for HIV-1 Permissiveness and Latent Infection.mBio. 2022 Dec 20;13(6):e0249622. doi: 10.1128/mbio.02496-22. Epub 2022 Oct 10. mBio. 2022. PMID: 36214569 Free PMC article.
-
CD38 Expression in a Subset of Memory T Cells Is Independent of Cell Cycling as a Correlate of HIV Disease Progression.Dis Markers. 2016;2016:9510756. doi: 10.1155/2016/9510756. Epub 2016 Mar 14. Dis Markers. 2016. PMID: 27064238 Free PMC article.
-
Elevated Basal Pre-infection CXCL10 in Plasma and in the Small Intestine after Infection Are Associated with More Rapid HIV/SIV Disease Onset.PLoS Pathog. 2016 Aug 10;12(8):e1005774. doi: 10.1371/journal.ppat.1005774. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27509048 Free PMC article.
-
Peripheral blood CD4+CCR6+ compartment differentiates HIV-1 infected or seropositive elite controllers from long-term successfully treated individuals.Commun Biol. 2022 Apr 13;5(1):357. doi: 10.1038/s42003-022-03315-x. Commun Biol. 2022. PMID: 35418589 Free PMC article.
References
-
- Douek DC, Picker LJ, Koup RA. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 2003;21:265–304. - PubMed
-
- Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 2002;53:557–593. - PubMed
-
- Geeraert L, Kraus G, Pomerantz RJ. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu. Rev. Med. 2008;59:487–501. - PubMed
-
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

