Identification and functional characterization of the first nucleobase transporter in mammals: implication in the species difference in the intestinal absorption mechanism of nucleobases and their analogs between higher primates and other mammals
- PMID: 20042597
- PMCID: PMC2825448
- DOI: 10.1074/jbc.M109.032961
Identification and functional characterization of the first nucleobase transporter in mammals: implication in the species difference in the intestinal absorption mechanism of nucleobases and their analogs between higher primates and other mammals
Abstract
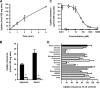
Nucleobases are important compounds that constitute nucleosides and nucleic acids. Although it has long been suggested that specific transporters are involved in their intestinal absorption and uptake in other tissues, none of their molecular entities have been identified in mammals to date. Here we describe identification of rat Slc23a4 as the first sodium-dependent nucleobase transporter (rSNBT1). The mRNA of rSNBT1 was expressed highly and only in the small intestine. When transiently expressed in HEK293 cells, rSNBT1 could transport uracil most efficiently. The transport of uracil mediated by rSNBT1 was sodium-dependent and saturable with a Michaelis constant of 21.2 microM. Thymine, guanine, hypoxanthine, and xanthine were also transported, but adenine was not. It was also suggested by studies of the inhibitory effect on rSNBT1-mediated uracil transport that several nucleobase analogs such as 5-fluorouracil are recognized by rSNBT1, but cytosine and nucleosides are not or only poorly recognized. Furthermore, rSNBT1 fused with green fluorescent protein was mainly localized at the apical membrane, when stably expressed in polarized Madin-Darby canine kidney II cells. These characteristics of rSNBT1 were almost fully in agreement with those of the carrier-mediated transport system involved in intestinal uracil uptake. Therefore, it is likely that rSNBT1 is its molecular entity or at least in part responsible for that. It was also found that the gene orthologous to the rSNBT1 gene is genetically defective in humans. This may have a biological and evolutional meaning in the transport and metabolism of nucleobases. The present study provides novel insights into the specific transport and metabolism of nucleobases and their analogs for therapeutic use.
Figures






Similar articles
-
Urate transport function of rat sodium-dependent nucleobase transporter 1.Physiol Rep. 2018 May;6(10):e13714. doi: 10.14814/phy2.13714. Physiol Rep. 2018. PMID: 29845779 Free PMC article.
-
Heterologous expression of the mammalian sodium-nucleobase transporter rSNBT1 in Leishmania tarentolae.Biochim Biophys Acta Biomembr. 2019 Sep 1;1861(9):1546-1557. doi: 10.1016/j.bbamem.2019.07.001. Epub 2019 Jul 5. Biochim Biophys Acta Biomembr. 2019. PMID: 31283918
-
Current Understanding of the Intestinal Absorption of Nucleobases and Analogs.Biol Pharm Bull. 2020;43(9):1293-1300. doi: 10.1248/bpb.b20-00342. Biol Pharm Bull. 2020. PMID: 32879202 Review.
-
Characterization of nucleobase transport by mouse Sertoli cell line TM4.Biol Pharm Bull. 2009 Mar;32(3):450-5. doi: 10.1248/bpb.32.450. Biol Pharm Bull. 2009. PMID: 19252294
-
Pyrimidine transport activities in trypanosomes.Trends Parasitol. 2007 May;23(5):187-9; discussion 190. doi: 10.1016/j.pt.2007.03.003. Epub 2007 Mar 19. Trends Parasitol. 2007. PMID: 17374509 Review.
Cited by
-
The Concise Guide to PHARMACOLOGY 2013/14: transporters.Br J Pharmacol. 2013 Dec;170(8):1706-96. doi: 10.1111/bph.12450. Br J Pharmacol. 2013. PMID: 24528242 Free PMC article.
-
Identification of agents that reduce renal hypoxia-reoxygenation injury using cell-based screening: purine nucleosides are alternative energy sources in LLC-PK1 cells during hypoxia.Arch Biochem Biophys. 2012 Jan 1;517(1):53-70. doi: 10.1016/j.abb.2011.11.005. Epub 2011 Nov 11. Arch Biochem Biophys. 2012. PMID: 22100704 Free PMC article.
-
A Targeted Metabolomics Assay to Measure Eight Purines in the Diet of Common Bottlenose Dolphins, Tursiops truncatus.J Chromatogr Sep Tech. 2016 Oct;7(5):334. doi: 10.4172/2157-7064.1000334. Epub 2016 Sep 19. J Chromatogr Sep Tech. 2016. PMID: 27904786 Free PMC article.
-
Impact of the loss of slc43a3 on 6-mercaptopurine absorption and tissue distribution in mice.Drug Metab Dispos. 2025 Apr;53(4):100054. doi: 10.1016/j.dmd.2025.100054. Epub 2025 Mar 3. Drug Metab Dispos. 2025. PMID: 40133022 Free PMC article.
-
Identification of the amino acid residue responsible for the myricetin sensitivity of human proton-coupled folate transporter.Sci Rep. 2019 Dec 2;9(1):18105. doi: 10.1038/s41598-019-54367-9. Sci Rep. 2019. PMID: 31792273 Free PMC article.
References
-
- Zöllner N. (1982) Proc. Nutr. Soc. 41, 329–342 - PubMed
-
- Burnstock G. (2007) Physiol. Rev. 87, 659–797 - PubMed
-
- Plagemann P. G., Wohlhueter R. M., Woffendin C. (1988) Biochim. Biophys. Acta 947, 405–443 - PubMed
-
- de Koning H., Diallinas G. (2000) Mol. Membr. Biol. 17, 75–94 - PubMed
-
- Gray J. H., Owen R. P., Giacomini K. M. (2004) Pflugers Arch. 447, 728–734 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

