Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3
- PMID: 20042598
- PMCID: PMC2825479
- DOI: 10.1074/jbc.M109.072405
Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3
Abstract
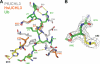
Like their human hosts, Plasmodium falciparum parasites rely on the ubiquitin-proteasome system for survival. We previously identified PfUCHL3, a deubiquitinating enzyme, and here we characterize its activity and changes in active site architecture upon binding to ubiquitin. We find strong evidence that PfUCHL3 is essential to parasite survival. The crystal structures of both PfUCHL3 alone and in complex with the ubiquitin-based suicide substrate UbVME suggest a rather rigid active site crossover loop that likely plays a role in restricting the size of ubiquitin adduct substrates. Molecular dynamics simulations of the structures and a model of the PfUCHL3-PfNedd8 complex allowed the identification of shared key interactions of ubiquitin and PfNedd8 with PfUCHL3, explaining the dual specificity of this enzyme. Distinct differences observed in ubiquitin binding between PfUCHL3 and its human counterpart make it likely that the parasitic DUB can be selectively targeted while leaving the human enzyme unaffected.
Figures


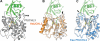
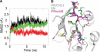
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

