TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication
- PMID: 20042610
- PMCID: PMC2844155
- DOI: 10.1074/jbc.M109.084038
TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication
Abstract
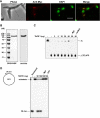
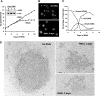
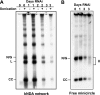
Kinetoplast DNA, the trypanosome mitochondrial genome, is a network of interlocked DNA rings including several thousand minicircles and a few dozen maxicircles. Minicircles replicate after release from the network, and their progeny reattach. Remarkably, trypanosomes have six mitochondrial DNA helicases related to yeast PIF1 helicase. Here we report that one of the six, TbPIF1, functions in minicircle replication. RNA interference (RNAi) of TbPIF1 causes a growth defect and kinetoplast DNA loss. Minicircle replication intermediates decrease during RNAi, and there is an accumulation of multiply interlocked, covalently closed minicircle dimers (fraction U). In studying the significance of fraction U, we found that this species also accumulates during RNAi of mitochondrial topoisomerase II. These data indicate that one function of TbPIF1 is an involvement, together with topoisomerase II, in the segregation of minicircle progeny.
Figures





Similar articles
-
A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei.Eukaryot Cell. 2011 Mar;10(3):445-54. doi: 10.1128/EC.00308-10. Epub 2011 Jan 21. Eukaryot Cell. 2011. PMID: 21257796 Free PMC article.
-
A new function of Trypanosoma brucei mitochondrial topoisomerase II is to maintain kinetoplast DNA network topology.Mol Microbiol. 2008 Dec;70(6):1465-76. doi: 10.1111/j.1365-2958.2008.06493.x. Epub 2008 Oct 22. Mol Microbiol. 2008. PMID: 19019151 Free PMC article.
-
Role of p38 in replication of Trypanosoma brucei kinetoplast DNA.Mol Cell Biol. 2006 Jul;26(14):5382-93. doi: 10.1128/MCB.00369-06. Mol Cell Biol. 2006. PMID: 16809774 Free PMC article.
-
Mitochondrial genome maintenance-the kinetoplast story.FEMS Microbiol Rev. 2023 Nov 1;47(6):fuac047. doi: 10.1093/femsre/fuac047. FEMS Microbiol Rev. 2023. PMID: 36449697 Free PMC article. Review.
-
The structure and replication of kinetoplast DNA.Annu Rev Microbiol. 1995;49:117-43. doi: 10.1146/annurev.mi.49.100195.001001. Annu Rev Microbiol. 1995. PMID: 8561456 Review.
Cited by
-
RNA interference screen reveals a high proportion of mitochondrial proteins essential for correct cell cycle progress in Trypanosoma brucei.BMC Genomics. 2015 Apr 15;16(1):297. doi: 10.1186/s12864-015-1505-5. BMC Genomics. 2015. PMID: 25888089 Free PMC article.
-
A passion for parasites.J Biol Chem. 2014 Dec 5;289(49):33712-29. doi: 10.1074/jbc.X114.620666. Epub 2014 Oct 21. J Biol Chem. 2014. PMID: 25336639 Free PMC article.
-
A leucine aminopeptidase is involved in kinetoplast DNA segregation in Trypanosoma brucei.PLoS Pathog. 2017 Apr 7;13(4):e1006310. doi: 10.1371/journal.ppat.1006310. eCollection 2017 Apr. PLoS Pathog. 2017. PMID: 28388690 Free PMC article.
-
Getting it done at the ends: Pif1 family DNA helicases and telomeres.DNA Repair (Amst). 2016 Aug;44:151-158. doi: 10.1016/j.dnarep.2016.05.021. Epub 2016 May 16. DNA Repair (Amst). 2016. PMID: 27233114 Free PMC article. Review.
-
A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei.Eukaryot Cell. 2011 Mar;10(3):445-54. doi: 10.1128/EC.00308-10. Epub 2011 Jan 21. Eukaryot Cell. 2011. PMID: 21257796 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

