Proof of principle for successful characterization of methicillin-resistant coagulase-negative staphylococci isolated from skin by use of Raman spectroscopy and pulsed-field gel electrophoresis
- PMID: 20042618
- PMCID: PMC2832465
- DOI: 10.1128/JCM.01153-09
Proof of principle for successful characterization of methicillin-resistant coagulase-negative staphylococci isolated from skin by use of Raman spectroscopy and pulsed-field gel electrophoresis
Abstract
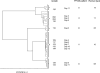
Coagulase-negative staphylococci (CNS) are among the most frequently isolated bacterial species in clinical microbiology, and most CNS-related infections are hospital acquired. Distinguishing between these frequently multiple-antibiotic-resistant isolates is important for both treatment and transmission control. In this study we used isolates of methicillin-resistant coagulase-negative staphylococci (MR-CNS) that were selected from a large surveillance study of the direct spread of MR-CNS. This strain collection was used to evaluate (i) Raman spectroscopy as a typing tool for MR-CNS isolates and (ii) diversity between colonies with identical and different morphologies. Reproducibility was high, with 215 of 216 (99.5%) of the replicate samples for 72 isolates ending up in the same cluster. The concordance with pulsed-field gel electrophoresis (PFGE)-based clusters was 94.4%. We also confirm that the skin of patients can be colonized with multiple MR-CNS types at the same time. Morphological differences between colonies from a single patient sample correlated with differences in Raman and PFGE types. Some morphologically indistinguishable colonies revealed different Raman and PFGE types. This indicates that multiple MR-CNS colonies should be examined to obtain a complete insight into the prevalence of different types and to be able to perform an accurate transmission analysis. Here we show that Raman spectroscopy is a reproducible typing system for MR-CNS isolates. It is a tool for screening variability within a collection of isolates. Because of the high throughput, it enables the analysis of multiple colonies per patient, which will enhance the quality of clinical and epidemiological studies.
Figures

References
-
- Ben Saida, N., A. Ferjeni, N. Benhadjtaher, K. Monastiri, and J. Boukadida. 2006. Clonality of clinical methicillin-resistant Staphylococcus epidermidis isolates in a neonatal intensive care unit. Pathol. Biol. (Paris) 54:337-342. - PubMed
-
- Casey, A. L., T. Worthington, P. A. Lambert, and T. S. Elliott. 2007. Evaluation of routine microbiological techniques for establishing the diagnosis of catheter-related bloodstream infection caused by coagulase-negative staphylococci. J. Med. Microbiol. 56:172-176. - PubMed
-
- Cookson, B. D., D. A. Robinson, A. B. Monk, S. Murchan, A. Deplano, R. de Ryck, M. J. Struelens, C. Scheel, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, C. Cuny, W. Witte, P. T. Tassios, N. J. Legakis, W. van Leeeuwen, A. van Belkum, A. Vindel, J. Garaizer, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, M. Muller-Premru, W. Hryniewicz, A. Rossney, B. O'Connell, B. D. Short, J. Thomas, S. O'Hanlon, and M. C. Enright. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus (MRSA) strains: the HARMONY collection. J. Clin. Microbiol. 45:1830-1837. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

