Multilocus sequence typing of Clostridium difficile
- PMID: 20042623
- PMCID: PMC2832416
- DOI: 10.1128/JCM.01796-09
Multilocus sequence typing of Clostridium difficile
Abstract
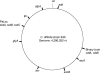
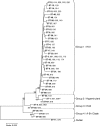
A robust high-throughput multilocus sequence typing (MLST) scheme for Clostridium difficile was developed and validated using a diverse collection of 50 reference isolates representing 45 different PCR ribotypes and 102 isolates from recent clinical samples. A total of 49 PCR ribotypes were represented overall. All isolates were typed by MLST and yielded 40 sequence types (STs). A web-accessible database was set up (http://pubmlst.org/cdifficile/) to facilitate the dissemination and comparison of C. difficile MLST genotyping data among laboratories. MLST and PCR ribotyping were similar in discriminatory abilities, having indices of discrimination of 0.90 and 0.92, respectively. Some STs corresponded to a single PCR ribotype (32/40), other STs corresponded to multiple PCR ribotypes (8/40), and, conversely, the PCR ribotype was not always predictive of the ST. The total number of variable nucleotide sites in the concatenated MLST sequences was 103/3,501 (2.9%). Concatenated MLST sequences were used to construct a neighbor-joining tree which identified four phylogenetic groups of STs and one outlier (ST-11; PCR ribotype 078). These groups apparently correlate with clades identified previously by comparative genomics. The MLST scheme was sufficiently robust to allow direct genotyping of C. difficile in total stool DNA extracts without isolate culture. The direct (nonculture) MLST approach may prove useful as a rapid genotyping method, potentially benefiting individual patients and informing hospital infection control.
Figures

References
-
- Bartlett, J. G. 2002. Clinical practice. Antibiotic-associated diarrhea. N. Engl. J. Med. 31:334-339. - PubMed
-
- Bartlett, J. G., and D. N. Gerding. 2008. Clinical recognition and diagnosis of Clostridium difficile infection. Clin. Infect. Dis. 46:S12-S18. - PubMed
-
- Braun, V., T. Hundsberger, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29-38. - PubMed
-
- Clabots, C. R., S. Johnson, K. M. Bettin, P. A. Mathie, M. E. Mulligan, D. R. Schaberg, L. R. Peterson, and D. N. Gerding. 1993. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J. Clin. Microbiol. 31:1870-1875. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

