Aerobic nonylphenol degradation and nitro-nonylphenol formation by microbial cultures from sediments
- PMID: 20043151
- PMCID: PMC2825322
- DOI: 10.1007/s00253-009-2394-9
Aerobic nonylphenol degradation and nitro-nonylphenol formation by microbial cultures from sediments
Abstract
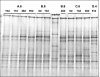
Nonylphenol (NP) is an estrogenic pollutant which is widely present in the aquatic environment. Biodegradation of NP can reduce the toxicological risk. In this study, aerobic biodegradation of NP in river sediment was investigated. The sediment used for the microcosm experiments was aged polluted with NP. The biodegradation of NP in the sediment occurred within 8 days with a lag phase of 2 days at 30 degrees C. During the biodegradation, nitro-nonylphenol metabolites were formed, which were further degraded to unknown compounds. The attached nitro-group originated from the ammonium in the medium. Five subsequent transfers were performed from original sediment and yielded a final stable population. In this NP-degrading culture, the microorganisms possibly involved in the biotransformation of NP to nitro-nonylphenol were related to ammonium-oxidizing bacteria. Besides the degradation of NP via nitro-nonylphenol, bacteria related to phenol-degrading species, which degrade phenol via ring cleavage, are abundantly present.
Figures






References
-
- Corvini PFX, Hollender J, Schumacher S, Prell J, Hommes G, Priefer U, Vinken R, Schaffer A. The degradation of alpha-quaternary nonylphenol isomers by Sphingomonas sp. strain TTNP3 involves a type II ipso-substitution mechanism. Appl Microbiol Biotechnol. 2006;70:114–122. doi: 10.1007/s00253-005-0080-0. - DOI - PubMed
-
- Corvini PFX, Meesters RJW, Mundt M, Schäffer A, Schmidt B, Schröder HF, Verstraete W, Vinken R, Hollender J. Contribution to the detection and identification of oxidation metabolites of nonylphenol in Sphingomonas sp. strain TTNP3. Biodegradation. 2007;18:233–245. doi: 10.1007/s10532-006-9058-6. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

